
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,325,506 – 6,325,611 |
| Length | 105 |
| Max. P | 0.935473 |

| Location | 6,325,506 – 6,325,611 |
|---|---|
| Length | 105 |
| Sequences | 15 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.18 |
| Shannon entropy | 0.49662 |
| G+C content | 0.55739 |
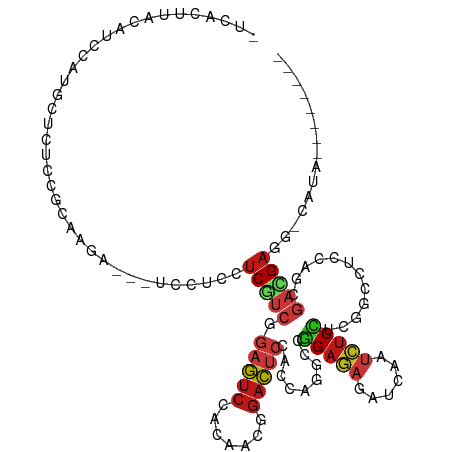
| Mean single sequence MFE | -33.95 |
| Consensus MFE | -14.14 |
| Energy contribution | -14.27 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.935473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
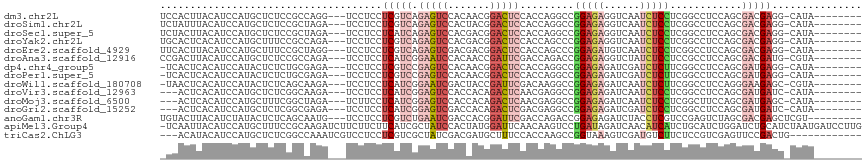
>dm3.chr2L 6325506 105 - 23011544 UCCACUUACAUCCAUGCUCUCCGCCAGG---UCCUCCUCGUCAGAGUCCACAACGGACUCCACCAGGCCGGAGAGGUCAAUCUCCUCGGCCUCCAGCGACGAGG-CAUA-------- .............((((...((....))---.....((((((.((((((.....))))))....((((((..((((....))))..)))))).....)))))))-))).-------- ( -40.70, z-score = -3.83, R) >droSim1.chr2L 6130437 105 - 22036055 UCUAUUUACAUCCAUGCUCUCCGCUAGA---UCCUCCUCGUCAGAGUCCACUACGGACUCCACCAGGCCGGAGAGGUCAAUCUCCUCGGCCUCCAGCGACGAGG-CAUA-------- .........(((...((.....))..))---)...(((((((.((((((.....))))))....((((((..((((....))))..)))))).....)))))))-....-------- ( -39.60, z-score = -3.71, R) >droSec1.super_5 4396294 105 - 5866729 UCUACUUACAUCCAUGCUCUCCGCUAGA---UCCUCCUCAUCAGAGUCCACGACGGACUCCACCAGGCCGGAGAGGUCAAUCUCCUCGGCCUCCAGCGACGAGG-CAUA-------- .............((((.(((((((.((---............((((((.....)))))).....(((((..((((....))))..))))))).))))..))))-))).-------- ( -38.40, z-score = -3.19, R) >droYak2.chr2L 15745201 105 + 22324452 UGCACUCACAUCCAUGCUUUCCGCCAGA---UCCUCCUCGUCAGAGUCCACGACGGACUCCACCAGCCCGGAGAGGUCAAUCUCCUCGGCCUCCAGCGACGAGG-CAUA-------- (((......(((...((.....))..))---)....((((((.((((((.....)))))).....(((.(((((......)))))..))).......)))))))-))..-------- ( -34.50, z-score = -1.99, R) >droEre2.scaffold_4929 15253838 105 - 26641161 UUCACUUACAUCCAUGCUUUCCGCUAGG---UCCUCCUCGUCAGAGUCGACGACGGACUCCACCAGCCCGGAGAUGUCAAUCUCCUCGGCCUCCAGCGACGAGG-CAUA-------- .............(((((((.((((.((---(..((((((((......))))).)))....))).(((.((((((....))))))..)))....))))..))))-))).-------- ( -37.30, z-score = -2.91, R) >droAna3.scaffold_12916 8270815 105 + 16180835 CCGACUUACAUCCAUGCUCUCCGCCAGA---UCCUCCUCAUCGGAAUCCACAACCGAUUCGACCAGACCGGAGAGGUCUAUCUCCUCCGCCUCCAGCGACGAUG-CGUA-------- .............((((((......(((---((((((..(((((.........)))))((.....))..)))).)))))........(((.....)))..)).)-))).-------- ( -22.00, z-score = 0.49, R) >dp4.chr4_group5 329502 104 + 2436548 -UCACUCACAUCCAUACUCUCUGCGAGA---UCCUCCUCGUCCGAGUCCACAACGGACUCCACCAGGCCGGAGAGAUCGAUCUCUUCGGCCUCCAGCGAUGAGG-CAUA-------- -...(((.((...........)).))).---....((((((((((((((.....))))))....((((((((((((....))))))))))))...).)))))))-....-------- ( -42.90, z-score = -4.34, R) >droPer1.super_5 325753 104 + 6813705 -UCACUCACAUCCAUACUCUCUGCGAGA---UCCUCCUCGUCCGAGUCCACAACGGACUCCACCAGGCCGGAGAGAUCGAUCUCUUCGGCCUCCAGCGAUGAGG-CAUA-------- -...(((.((...........)).))).---....((((((((((((((.....))))))....((((((((((((....))))))))))))...).)))))))-....-------- ( -42.90, z-score = -4.34, R) >droWil1.scaffold_180708 5354984 104 + 12563649 -UAACUCACAUCCAUACUCUCAGCAAGA---UCCUCCUCAUCGGAAUCGACUACCGAUUCGACAAGGCCGGAGAGAUCAAUCUCUUCGGCCUCCAGGGAAGAGC-CGUA-------- -...(((..........(((.....)))---((((........((((((.....))))))....((((((((((((....))))))))))))...)))).))).-....-------- ( -33.40, z-score = -2.52, R) >droVir3.scaffold_12963 17394220 102 + 20206255 ---ACUCACAUCCAUGCUCUCGGCAAGA---UCCUCCUCAUCGGAGUCCACCACAGACUCAACGAGGCCGGAGAGAUCAAUCUCCUCGGCCUCCAGCGAUGAUC-CAUA-------- ---...........(((.....)))...---......(((((((((((.......)))))...(((((((..((((....))))..)))))))...))))))..-....-------- ( -36.80, z-score = -3.47, R) >droMoj3.scaffold_6500 19278392 102 + 32352404 ---ACUCACAUCCAUGCUUUCGGCUAGA---UCUUCCUCAUCGGAGUCCACCACAGACUCAACGAGGCCGGAGAGAUCAAUCUCCUCGGCUUCCAGCGAUGAGC-CAUA-------- ---......(((...((.....))..))---)....((((((((((((.......)))))...(((((((..((((....))))..)))))))...))))))).-....-------- ( -35.80, z-score = -3.22, R) >droGri2.scaffold_15252 14757664 102 + 17193109 ---ACUCACAUCCAUGCUCUCGGCGAGA---UCCUCCUCAUCGGAGUCGACCACAGACUCGACGAGGCCGGAGAGAUCGAUCUCCUCGGCCUCCAGCGAUGAUC-CAUA-------- ---.(((....((........)).))).---......(((((((((((.......)))))...(((((((..((((....))))..)))))))...))))))..-....-------- ( -37.70, z-score = -1.41, R) >anoGam1.chr3R 30396396 105 - 53272125 UGUACUUACAUCUAUACUCUCAGCAAUG---UCCUCCUCGUCUGAAUCGACCACGGAUUCGACCAGACCGGAGAGAUCUACCUCGUCCGAGUCUAGCGACGAGCUCGU--------- ............................---.....((((((((((((.......))))))...(((((((((((......))).)))).))))...)))))).....--------- ( -27.10, z-score = 0.11, R) >apiMel3.Group4 4257953 116 + 10796202 -UCAAUUACAUCCAUGCUUUCCGCAAGAUCUUCUUCUUCAUCGCUAUCCACUAUGGAUUCAACAAGUCCUGAUAGAUCAACAUCAUCUGCAUCUGGAUCUGCAUCUAAUGAUCCUUG -.............(((.....))).(((((((............((((.....))))............)).)))))................(((((..........)))))... ( -17.45, z-score = 0.07, R) >triCas2.ChLG3 27733031 102 - 32080666 ---ACAUACAUCCAUGCUCUCGGCCAAAUCGUCCUCCUCGUCGCUAUCGACGAUGCUUUCCACCAAGCCGGUAAAGUCGAUGUCUUCUCCGUCGAGUUCCGACUG------------ ---....(((((.((....(((((.............((((((....)))))).............)))))....)).))))).......((((.....))))..------------ ( -22.77, z-score = -1.01, R) >consensus _UCACUUACAUCCAUGCUCUCCGCAAGA___UCCUCCUCGUCGGAGUCCACAACGGACUCCACCAGGCCGGAGAGAUCAAUCUCCUCGGCCUCCAGCGACGAGG_CAUA________ .....................................(((((.(((((.......))))).........(((((......)))))............)))))............... (-14.14 = -14.27 + 0.13)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:21:03 2011