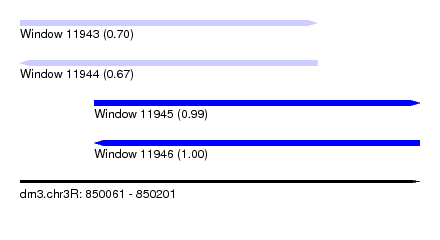
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 850,061 – 850,201 |
| Length | 140 |
| Max. P | 0.998779 |

| Location | 850,061 – 850,165 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 89.83 |
| Shannon entropy | 0.20830 |
| G+C content | 0.39110 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -17.11 |
| Energy contribution | -16.79 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.699149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
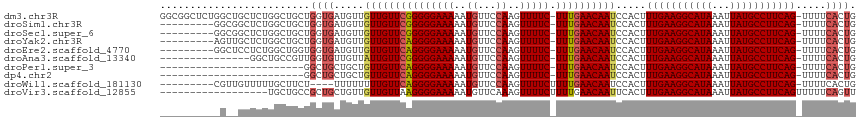
>dm3.chr3R 850061 104 + 27905053 ------ACAGCUGACAGCC---AACAACUCAUGCACA----GUGAAAACUGAAGGCAUAAUUUAUGCCUUCAAAGUGGAUUGUUCAAA-GAAAACUUGGAACAUUUUUCCCCCGAACA ------...(((((.....---......))).))..(----((....)))(((((((((...))))))))).....(((.(((((.((-(....))).)))))....)))........ ( -24.10, z-score = -1.66, R) >droSim1.chr3R 884974 104 + 27517382 ------ACAGCUGACAGCC---AACAACUCAUGCACA----GUGAAAACUGAAGGCAUAAUUUAUGCCUUCAAAGUGGAUUGUUCAAA-GAAAACUUGGAACAUUUUUCCCCCGAACA ------...(((((.....---......))).))..(----((....)))(((((((((...))))))))).....(((.(((((.((-(....))).)))))....)))........ ( -24.10, z-score = -1.66, R) >droSec1.super_6 986843 104 + 4358794 ------ACAGCUGACAGCC---AACAACUCAUGCACA----GUGAAAACUGAAGGCAUAAUUUAUGCCUUCAAAGUGGAUUGUUCAAA-GAAAACUUGGAACAUUUUUCCCCCGAACA ------...(((((.....---......))).))..(----((....)))(((((((((...))))))))).....(((.(((((.((-(....))).)))))....)))........ ( -24.10, z-score = -1.66, R) >droYak2.chr3R 1191046 104 + 28832112 ------ACAGCUGACAGCC---AACAGCUCAUGCACA----GUGAAAACUGAAGGCAUAAUUUAUGCCUUCAAAGUGGAUUGUUCAAA-GAAAACUUGGAACAUUUUUCCCCUGAACA ------..(((((......---..))))).((.((((----((....)))(((((((((...)))))))))...))).))((((((((-(....)))((((.....))))..)))))) ( -29.10, z-score = -2.67, R) >droEre2.scaffold_4770 1138200 104 + 17746568 ------ACAGCUGACAGCC---AACAGCUCAUGCACA----GUGAAAACUGAAGGCAUAAUUUAUGCCUUCAAAGUGGAUUGUUCAAA-GAAAACUUGGAACAUUUUUCCCCUGAACA ------..(((((......---..))))).((.((((----((....)))(((((((((...)))))))))...))).))((((((((-(....)))((((.....))))..)))))) ( -29.10, z-score = -2.67, R) >droAna3.scaffold_13340 23059502 104 + 23697760 ------ACAGCUGACAGCC---AACAACUCAUGCACA----GUGAAAACUGAAGGCAUAAUUUAUGCCUUCAAAGUGGAUUGUUCAAA-GAAAACUUGGAACAUUUUUCCCCCGAACA ------...(((((.....---......))).))..(----((....)))(((((((((...))))))))).....(((.(((((.((-(....))).)))))....)))........ ( -24.10, z-score = -1.66, R) >droPer1.super_3 2636394 108 - 7375914 ------ACAGCUGACAGCC---AACAACUCAUGCACACACAGUGAAAACUGAAGGCAUAAUUUAUGCCUUCAAAGUGGAUUGUUCAAA-GAAAACUUGGAACAUUUUUCCCCUGAACA ------.(((..((.....---........((.(((....(((....)))(((((((((...)))))))))...))).))(((((.((-(....))).)))))....))..))).... ( -25.70, z-score = -1.96, R) >dp4.chr2 8835995 108 - 30794189 ------ACAGCUGACAGCC---AACAACUCAUGCACACACAGUGAAAACUGAAGGCAUAAUUUAUGCCUUCAAAGUGGAUUGUUCAAA-GAAAACUUGGAACAUUUUUCCCCUGAACA ------.(((..((.....---........((.(((....(((....)))(((((((((...)))))))))...))).))(((((.((-(....))).)))))....))..))).... ( -25.70, z-score = -1.96, R) >droWil1.scaffold_181130 3571349 106 - 16660200 ------ACAGCUGACAGCCC--AACAACUCAUGCACA----GUGAAAACUGAAGGCAUAAUUUAUGCCUUCAAAGUGGAUUGUUCAAAAGAAAACUUGGAACAUUUUUCCCCUGAACA ------.(((..((((..((--(.....((((.....----))))....((((((((((...))))))))))...)))..)))).............((((.....)))).))).... ( -24.60, z-score = -1.61, R) >droVir3.scaffold_12855 2130178 102 - 10161210 -------------ACAGCCAACAACAAUUCAUUCGAAC---UGAAAAACUGAAGGCAUAAUUUAUGCCUUCAAAGUGAAUUGUUCAAAAGAAAACUUUGAACAUUUUUCCCCUUAACA -------------...........((.(((....))).---))......((((((((((...))))))))))(((.(((.((((((((.......))))))))...)))..))).... ( -22.10, z-score = -3.66, R) >droMoj3.scaffold_6540 10879546 115 - 34148556 AGGCGACAAGCUGACAGCCAACAGCAAUUCAUGCGAAC---UGAAAAACUGAAGGCAUAAUUUAUGCCUUCAAAGUGAAUUGUUCAAAAGAAAACUUGGAACAUUUUUCCCCUUAACA .(((............)))...((((((((((......---(....)..((((((((((...))))))))))..)))))))))).............((((.....))))........ ( -26.50, z-score = -1.94, R) >droGri2.scaffold_15095 39289 105 - 78014 ------CAAGCUGACAGGCAAGAACAAUUCAUGCACAG---UGCAAAACUGAAGGCAUAAUUUAUGCCUUCAAAGUGAAUUGUUCAAA-GAAAACUUGGAACAUUUUUCCCCUUA--- ------...(((....)))..(((((((((((((....---.)).....((((((((((...))))))))))..)))))))))))...-........((((.....)))).....--- ( -33.00, z-score = -4.33, R) >consensus ______ACAGCUGACAGCC___AACAACUCAUGCACA____GUGAAAACUGAAGGCAUAAUUUAUGCCUUCAAAGUGGAUUGUUCAAA_GAAAACUUGGAACAUUUUUCCCCUGAACA ......................(((((.((((.................((((((((((...))))))))))..)))).))))).............((((.....))))........ (-17.11 = -16.79 + -0.32)
| Location | 850,061 – 850,165 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 89.83 |
| Shannon entropy | 0.20830 |
| G+C content | 0.39110 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -20.50 |
| Energy contribution | -20.74 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.671672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 850061 104 - 27905053 UGUUCGGGGGAAAAAUGUUCCAAGUUUUC-UUUGAACAAUCCACUUUGAAGGCAUAAAUUAUGCCUUCAGUUUUCAC----UGUGCAUGAGUUGUU---GGCUGUCAGCUGU------ (((.(((..(((((.(((((.(((....)-)).))))).......(((((((((((...)))))))))))))))).)----)).)))..(((((..---......)))))..------ ( -30.90, z-score = -2.12, R) >droSim1.chr3R 884974 104 - 27517382 UGUUCGGGGGAAAAAUGUUCCAAGUUUUC-UUUGAACAAUCCACUUUGAAGGCAUAAAUUAUGCCUUCAGUUUUCAC----UGUGCAUGAGUUGUU---GGCUGUCAGCUGU------ (((.(((..(((((.(((((.(((....)-)).))))).......(((((((((((...)))))))))))))))).)----)).)))..(((((..---......)))))..------ ( -30.90, z-score = -2.12, R) >droSec1.super_6 986843 104 - 4358794 UGUUCGGGGGAAAAAUGUUCCAAGUUUUC-UUUGAACAAUCCACUUUGAAGGCAUAAAUUAUGCCUUCAGUUUUCAC----UGUGCAUGAGUUGUU---GGCUGUCAGCUGU------ (((.(((..(((((.(((((.(((....)-)).))))).......(((((((((((...)))))))))))))))).)----)).)))..(((((..---......)))))..------ ( -30.90, z-score = -2.12, R) >droYak2.chr3R 1191046 104 - 28832112 UGUUCAGGGGAAAAAUGUUCCAAGUUUUC-UUUGAACAAUCCACUUUGAAGGCAUAAAUUAUGCCUUCAGUUUUCAC----UGUGCAUGAGCUGUU---GGCUGUCAGCUGU------ (((.(((..(((((.(((((.(((....)-)).))))).......(((((((((((...)))))))))))))))).)----)).)))..(((((..---......)))))..------ ( -34.00, z-score = -2.90, R) >droEre2.scaffold_4770 1138200 104 - 17746568 UGUUCAGGGGAAAAAUGUUCCAAGUUUUC-UUUGAACAAUCCACUUUGAAGGCAUAAAUUAUGCCUUCAGUUUUCAC----UGUGCAUGAGCUGUU---GGCUGUCAGCUGU------ (((.(((..(((((.(((((.(((....)-)).))))).......(((((((((((...)))))))))))))))).)----)).)))..(((((..---......)))))..------ ( -34.00, z-score = -2.90, R) >droAna3.scaffold_13340 23059502 104 - 23697760 UGUUCGGGGGAAAAAUGUUCCAAGUUUUC-UUUGAACAAUCCACUUUGAAGGCAUAAAUUAUGCCUUCAGUUUUCAC----UGUGCAUGAGUUGUU---GGCUGUCAGCUGU------ (((.(((..(((((.(((((.(((....)-)).))))).......(((((((((((...)))))))))))))))).)----)).)))..(((((..---......)))))..------ ( -30.90, z-score = -2.12, R) >droPer1.super_3 2636394 108 + 7375914 UGUUCAGGGGAAAAAUGUUCCAAGUUUUC-UUUGAACAAUCCACUUUGAAGGCAUAAAUUAUGCCUUCAGUUUUCACUGUGUGUGCAUGAGUUGUU---GGCUGUCAGCUGU------ (((((((((((((..((...))..)))))-)))))))).......(((((((((((...)))))))))))....(((.....)))....(((((..---......)))))..------ ( -31.10, z-score = -1.77, R) >dp4.chr2 8835995 108 + 30794189 UGUUCAGGGGAAAAAUGUUCCAAGUUUUC-UUUGAACAAUCCACUUUGAAGGCAUAAAUUAUGCCUUCAGUUUUCACUGUGUGUGCAUGAGUUGUU---GGCUGUCAGCUGU------ (((((((((((((..((...))..)))))-)))))))).......(((((((((((...)))))))))))....(((.....)))....(((((..---......)))))..------ ( -31.10, z-score = -1.77, R) >droWil1.scaffold_181130 3571349 106 + 16660200 UGUUCAGGGGAAAAAUGUUCCAAGUUUUCUUUUGAACAAUCCACUUUGAAGGCAUAAAUUAUGCCUUCAGUUUUCAC----UGUGCAUGAGUUGUU--GGGCUGUCAGCUGU------ ((((((((((((((.(......).)))))))))))))).........(((((((((...)))))))))((((..(((----(..(((.....))).--.)).))..))))..------ ( -30.20, z-score = -1.60, R) >droVir3.scaffold_12855 2130178 102 + 10161210 UGUUAAGGGGAAAAAUGUUCAAAGUUUUCUUUUGAACAAUUCACUUUGAAGGCAUAAAUUAUGCCUUCAGUUUUUCA---GUUCGAAUGAAUUGUUGUUGGCUGU------------- .(((((.(((((((((((((((((.....)))))))).........((((((((((...)))))))))))))))))(---((((....))))).)).)))))...------------- ( -28.60, z-score = -3.17, R) >droMoj3.scaffold_6540 10879546 115 + 34148556 UGUUAAGGGGAAAAAUGUUCCAAGUUUUCUUUUGAACAAUUCACUUUGAAGGCAUAAAUUAUGCCUUCAGUUUUUCA---GUUCGCAUGAAUUGCUGUUGGCUGUCAGCUUGUCGCCU ......((((((((((((((.(((....)))..)))).........((((((((((...))))))))))))))))).---........((...((((........))))...)).))) ( -29.90, z-score = -1.46, R) >droGri2.scaffold_15095 39289 105 + 78014 ---UAAGGGGAAAAAUGUUCCAAGUUUUC-UUUGAACAAUUCACUUUGAAGGCAUAAAUUAUGCCUUCAGUUUUGCA---CUGUGCAUGAAUUGUUCUUGCCUGUCAGCUUG------ ---..(((((((.....))))........-...((((((((((..(((((((((((...)))))))))))...(((.---....)))))))))))))...))).........------ ( -33.10, z-score = -3.57, R) >consensus UGUUCAGGGGAAAAAUGUUCCAAGUUUUC_UUUGAACAAUCCACUUUGAAGGCAUAAAUUAUGCCUUCAGUUUUCAC____UGUGCAUGAGUUGUU___GGCUGUCAGCUGU______ ......((((((.....))))..((((......))))...))...(((((((((((...)))))))))))..............((.(((...((.....))..)))))......... (-20.50 = -20.74 + 0.24)

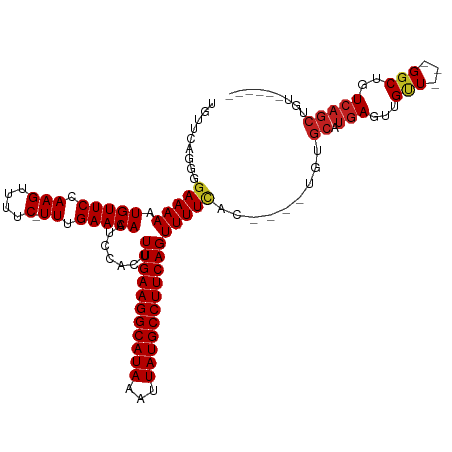
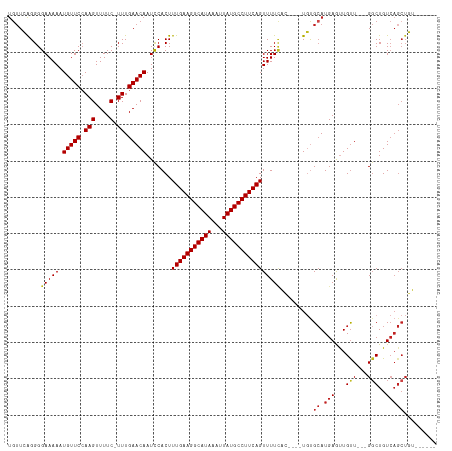
| Location | 850,087 – 850,201 |
|---|---|
| Length | 114 |
| Sequences | 10 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 84.77 |
| Shannon entropy | 0.30805 |
| G+C content | 0.40459 |
| Mean single sequence MFE | -25.82 |
| Consensus MFE | -18.82 |
| Energy contribution | -19.24 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.989047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 850087 114 + 27905053 CAGUGAAAA-CUGAAGGCAUAAUUUAUGCCUUCAAAGUGGAUUGUUCAAA-GAAAACUUGGAACAUUUUUCCCCCGAACAACAACAUCACCAGCAGCCAGAGCAGCCAGAGCCGCC .(((....)-))(((((((((...)))))))))...((((.((((((.((-(....)))((((.....))))...))))))...........((.((....)).)).....)))). ( -28.40, z-score = -2.34, R) >droSim1.chr3R 885000 105 + 27517382 CAGUGAAAA-CUGAAGGCAUAAUUUAUGCCUUCAAAGUGGAUUGUUCAAA-GAAAACUUGGAACAUUUUUCCCCCGAACAACAACAUCACCAGCAGCCAGAGCCGCC--------- ..((((...-.((((((((((...))))))))))..((...((((((.((-(....)))((((.....))))...))))))..)).))))..((.((....)).)).--------- ( -27.80, z-score = -3.10, R) >droSec1.super_6 986869 105 + 4358794 CAGUGAAAA-CUGAAGGCAUAAUUUAUGCCUUCAAAGUGGAUUGUUCAAA-GAAAACUUGGAACAUUUUUCCCCCGAACAACAACAUCACCAGCAGCCAGAGCCGCC--------- ..((((...-.((((((((((...))))))))))..((...((((((.((-(....)))((((.....))))...))))))..)).))))..((.((....)).)).--------- ( -27.80, z-score = -3.10, R) >droYak2.chr3R 1191072 105 + 28832112 CAGUGAAAA-CUGAAGGCAUAAUUUAUGCCUUCAAAGUGGAUUGUUCAAA-GAAAACUUGGAACAUUUUUCCCCUGAACAACAACAUCACCAGCAGCCAGAGCAACU--------- ..((((...-.((((((((((...)))))))))).((.(((.(((((.((-(....))).)))))....))).))...........))))..((.......))....--------- ( -27.00, z-score = -2.66, R) >droEre2.scaffold_4770 1138226 105 + 17746568 CAGUGAAAA-CUGAAGGCAUAAUUUAUGCCUUCAAAGUGGAUUGUUCAAA-GAAAACUUGGAACAUUUUUCCCCUGAACAACAACAUCACCACCAGCCAGAGGAGCC--------- ..((((...-.((((((((((...)))))))))).((.(((.(((((.((-(....))).)))))....))).))...........))))..((.......))....--------- ( -26.60, z-score = -2.28, R) >droAna3.scaffold_13340 23059528 99 + 23697760 CAGUGAAAA-CUGAAGGCAUAAUUUAUGCCUUCAAAGUGGAUUGUUCAAA-GAAAACUUGGAACAUUUUUCCCCCGAACAAUAACAACACCAACGGCAGCC--------------- ..(((....-.((((((((((...))))))))))..((..(((((((.((-(....)))((((.....))))...))))))).))..)))...........--------------- ( -25.40, z-score = -2.92, R) >droPer1.super_3 2636424 90 - 7375914 CAGUGAAAA-CUGAAGGCAUAAUUUAUGCCUUCAAAGUGGAUUGUUCAAA-GAAAACUUGGAACAUUUUUCCCCUGAACAACAGCAGCAGCC------------------------ .(((....)-))(((((((((...)))))))))..((.(((.(((((.((-(....))).)))))....))).))........((....)).------------------------ ( -24.00, z-score = -2.35, R) >dp4.chr2 8836025 90 - 30794189 CAGUGAAAA-CUGAAGGCAUAAUUUAUGCCUUCAAAGUGGAUUGUUCAAA-GAAAACUUGGAACAUUUUUCCCCUGAACAACAGCAGCAGCC------------------------ .(((....)-))(((((((((...)))))))))..((.(((.(((((.((-(....))).)))))....))).))........((....)).------------------------ ( -24.00, z-score = -2.35, R) >droWil1.scaffold_181130 3571376 102 - 16660200 CAGUGAAAA-CUGAAGGCAUAAUUUAUGCCUUCAAAGUGGAUUGUUCAAAAGAAAACUUGGAACAUUUUUCCCCUGAACAAAAAAAA----AGAAGCAAAAACAACG--------- .(((....)-))(((((((((...)))))))))..((.(((.(((((..(((....))).)))))....))).))............----................--------- ( -22.30, z-score = -2.68, R) >droVir3.scaffold_12855 2130200 98 - 10161210 AACUGAAAAACUGAAGGCAUAAUUUAUGCCUUCAAAGUGAAUUGUUCAAAAGAAAACUUUGAACAUUUUUCCCCUUAACAACAACAGCAGCGGCAGCA------------------ ..(((......((((((((((...))))))))))(((.(((.((((((((.......))))))))...)))..)))........)))..((....)).------------------ ( -24.90, z-score = -3.17, R) >consensus CAGUGAAAA_CUGAAGGCAUAAUUUAUGCCUUCAAAGUGGAUUGUUCAAA_GAAAACUUGGAACAUUUUUCCCCUGAACAACAACAUCACCAGCAGCCAGAGC__C__________ ..(((......((((((((((...))))))))))..((...((((((....(((((..........)))))....))))))..))..))).......................... (-18.82 = -19.24 + 0.42)

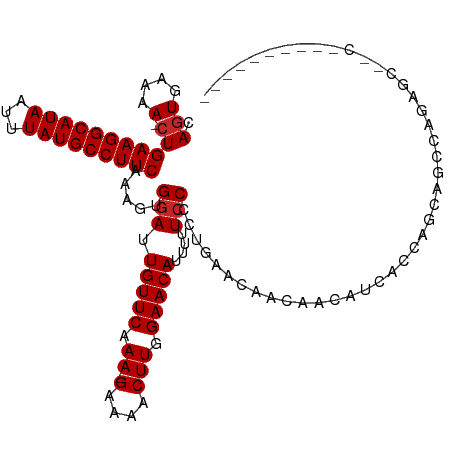
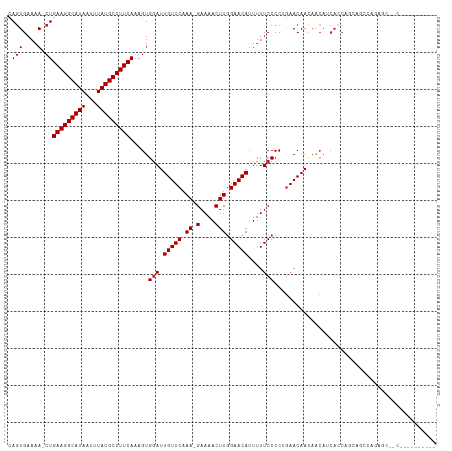
| Location | 850,087 – 850,201 |
|---|---|
| Length | 114 |
| Sequences | 10 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 84.77 |
| Shannon entropy | 0.30805 |
| G+C content | 0.40459 |
| Mean single sequence MFE | -33.79 |
| Consensus MFE | -24.27 |
| Energy contribution | -24.63 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.69 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.49 |
| SVM RNA-class probability | 0.998779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
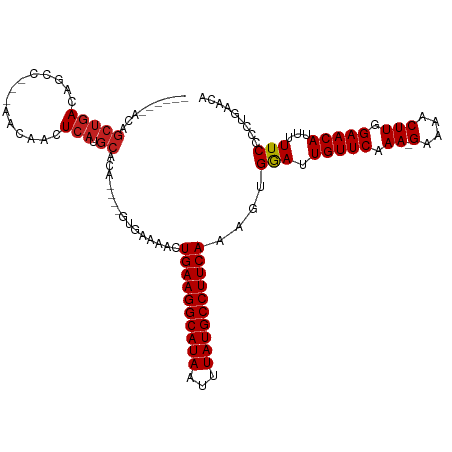
>dm3.chr3R 850087 114 - 27905053 GGCGGCUCUGGCUGCUCUGGCUGCUGGUGAUGUUGUUGUUCGGGGGAAAAAUGUUCCAAGUUUUC-UUUGAACAAUCCACUUUGAAGGCAUAAAUUAUGCCUUCAG-UUUUCACUG (((((((..((...))..)))))))(((((....(((((((((((((((..((...))..)))))-)))))))))).....(((((((((((...)))))))))))-...))))). ( -40.90, z-score = -3.73, R) >droSim1.chr3R 885000 105 - 27517382 ---------GGCGGCUCUGGCUGCUGGUGAUGUUGUUGUUCGGGGGAAAAAUGUUCCAAGUUUUC-UUUGAACAAUCCACUUUGAAGGCAUAAAUUAUGCCUUCAG-UUUUCACUG ---------((((((....))))))(((((....(((((((((((((((..((...))..)))))-)))))))))).....(((((((((((...)))))))))))-...))))). ( -40.80, z-score = -4.89, R) >droSec1.super_6 986869 105 - 4358794 ---------GGCGGCUCUGGCUGCUGGUGAUGUUGUUGUUCGGGGGAAAAAUGUUCCAAGUUUUC-UUUGAACAAUCCACUUUGAAGGCAUAAAUUAUGCCUUCAG-UUUUCACUG ---------((((((....))))))(((((....(((((((((((((((..((...))..)))))-)))))))))).....(((((((((((...)))))))))))-...))))). ( -40.80, z-score = -4.89, R) >droYak2.chr3R 1191072 105 - 28832112 ---------AGUUGCUCUGGCUGCUGGUGAUGUUGUUGUUCAGGGGAAAAAUGUUCCAAGUUUUC-UUUGAACAAUCCACUUUGAAGGCAUAAAUUAUGCCUUCAG-UUUUCACUG ---------(((.((....)).)))(((((....(((((((((((((((..((...))..)))))-)))))))))).....(((((((((((...)))))))))))-...))))). ( -36.10, z-score = -3.93, R) >droEre2.scaffold_4770 1138226 105 - 17746568 ---------GGCUCCUCUGGCUGGUGGUGAUGUUGUUGUUCAGGGGAAAAAUGUUCCAAGUUUUC-UUUGAACAAUCCACUUUGAAGGCAUAAAUUAUGCCUUCAG-UUUUCACUG ---------((((.....))))...(((((....(((((((((((((((..((...))..)))))-)))))))))).....(((((((((((...)))))))))))-...))))). ( -35.00, z-score = -3.28, R) >droAna3.scaffold_13340 23059528 99 - 23697760 ---------------GGCUGCCGUUGGUGUUGUUAUUGUUCGGGGGAAAAAUGUUCCAAGUUUUC-UUUGAACAAUCCACUUUGAAGGCAUAAAUUAUGCCUUCAG-UUUUCACUG ---------------..........((((.....(((((((((((((((..((...))..)))))-)))))))))).....(((((((((((...)))))))))))-....)))). ( -29.70, z-score = -2.74, R) >droPer1.super_3 2636424 90 + 7375914 ------------------------GGCUGCUGCUGUUGUUCAGGGGAAAAAUGUUCCAAGUUUUC-UUUGAACAAUCCACUUUGAAGGCAUAAAUUAUGCCUUCAG-UUUUCACUG ------------------------(((....)))(((((((((((((((..((...))..)))))-)))))))))).....(((((((((((...)))))))))))-......... ( -30.00, z-score = -3.70, R) >dp4.chr2 8836025 90 + 30794189 ------------------------GGCUGCUGCUGUUGUUCAGGGGAAAAAUGUUCCAAGUUUUC-UUUGAACAAUCCACUUUGAAGGCAUAAAUUAUGCCUUCAG-UUUUCACUG ------------------------(((....)))(((((((((((((((..((...))..)))))-)))))))))).....(((((((((((...)))))))))))-......... ( -30.00, z-score = -3.70, R) >droWil1.scaffold_181130 3571376 102 + 16660200 ---------CGUUGUUUUUGCUUCU----UUUUUUUUGUUCAGGGGAAAAAUGUUCCAAGUUUUCUUUUGAACAAUCCACUUUGAAGGCAUAAAUUAUGCCUUCAG-UUUUCACUG ---------................----......(((((((((((((((.(......).)))))))))))))))......(((((((((((...)))))))))))-......... ( -27.40, z-score = -3.66, R) >droVir3.scaffold_12855 2130200 98 + 10161210 ------------------UGCUGCCGCUGCUGUUGUUGUUAAGGGGAAAAAUGUUCAAAGUUUUCUUUUGAACAAUUCACUUUGAAGGCAUAAAUUAUGCCUUCAGUUUUUCAGUU ------------------.(((.((...((.......))...)).(((((((((((((((.....)))))))).........((((((((((...)))))))))))))))))))). ( -27.20, z-score = -2.35, R) >consensus __________G__GCUCUGGCUGCUGGUGAUGUUGUUGUUCAGGGGAAAAAUGUUCCAAGUUUUC_UUUGAACAAUCCACUUUGAAGGCAUAAAUUAUGCCUUCAG_UUUUCACUG .........................((((.....((((((((((.(((((.(......).))))).))))))))))......((((((((((...))))))))))......)))). (-24.27 = -24.63 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:50:31 2011