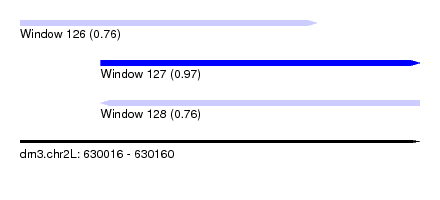
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 630,016 – 630,160 |
| Length | 144 |
| Max. P | 0.974208 |

| Location | 630,016 – 630,123 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 83.08 |
| Shannon entropy | 0.32173 |
| G+C content | 0.53244 |
| Mean single sequence MFE | -39.32 |
| Consensus MFE | -30.73 |
| Energy contribution | -30.77 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.758560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
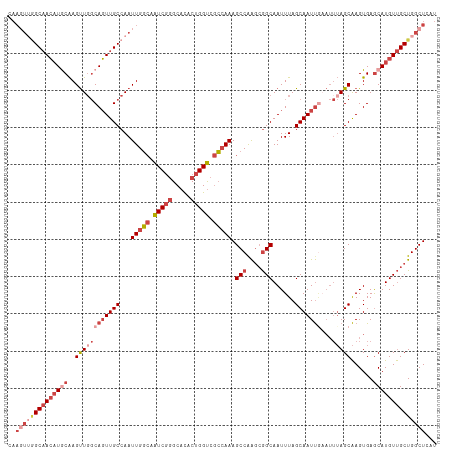
>dm3.chr2L 630016 107 + 23011544 CUGCAGGUGGC----AGUUUGGAGCCUUCAAUUCAAGUUGGCAACAUGCAAGUUGGCAGUUGCCAAUUGGCAAUCGGGCACACUGGUCGCCAAAGCCAAGCGGCAAUUUAG ((((...((((----..((((((........))))))((((((((.(((......)))))))))))(((((.(((((.....))))).))))).)))).))))........ ( -40.30, z-score = -1.93, R) >droSim1.chr2L 636500 107 + 22036055 CUGCAGGUGGC----AGUUUGGAGCCUUCAAUUCAAGUUGGCAACAUGCAAGUUGGCAGUUGCCAAUUGGCAAUCGGGCACACUGGUCGCCAAAGCCGAGCGGCAAUUUAG ((((...((((----..((((((........))))))((((((((.(((......)))))))))))(((((.(((((.....))))).))))).)))).))))........ ( -39.60, z-score = -1.57, R) >droSec1.super_14 612202 107 + 2068291 CUGCAGGUGGC----AGUUUGGUGCCUUCAACUCAAUUUGGCAACAUGCAAGUUUGCAGUUGCCAAUUGGCAAUCGGGCACACUGGUCGCCAAAGCCGAGCGGCAAUUUAG .....((((((----(((...((((((.....((((((.((((((.((((....)))))))))))))))).....))))))))).))))))...(((....)))....... ( -43.50, z-score = -2.69, R) >droYak2.chr2L 620002 107 + 22324452 CUGCAGGUGGC----AGCUUGGAGCCUUCAAUUCAAGUUGGCAACAUACAAGUUGGCAGUUGCCAAUUGGCAAUCGGGCACACUGGUCGCCAAAGCCAAGCGGCAAUUUAG ((((...((((----((((((((........))))))))((((((...(.....)...))))))..(((((.(((((.....))))).))))).)))).))))........ ( -39.00, z-score = -1.80, R) >droEre2.scaffold_4929 688004 107 + 26641161 CUGCAGGUGGC----AGUUUGGAGCCUUCAAUUCAAGUAAGCAACAUACAAGUUGGCAGUUGCCAAUUGCCAAUCGGGCACACUGGUCGGCAAAGCCAAGGGGCAAUUUAG .(((...((((----.((.((..(((((......)))...))..)).)).(((((((....)))))))(((.(((((.....))))).)))...))))....)))...... ( -32.20, z-score = -0.57, R) >droAna3.scaffold_12916 1624501 109 + 16180835 CUGCAGGUGGCCCGCAGUGGCAGGUUCCGAGCUGCUAGCUGCUAGUUGCGAGCUGCCACUUGCCAAUUGGAAGUCUCGC-UUCUGGCC-CAAAAGCCAGCCAGCAAUUUAG ..(((((((((..((((((((((.(....).)))))).)))).(((.....))))))))))))..............((-(.(((((.-.....)))))..)))....... ( -41.30, z-score = 0.01, R) >consensus CUGCAGGUGGC____AGUUUGGAGCCUUCAAUUCAAGUUGGCAACAUGCAAGUUGGCAGUUGCCAAUUGGCAAUCGGGCACACUGGUCGCCAAAGCCAAGCGGCAAUUUAG .(((...((((......((((((........))))))((((((((.(((......)))))))))))(((((.(((((.....))))).))))).))))....)))...... (-30.73 = -30.77 + 0.04)

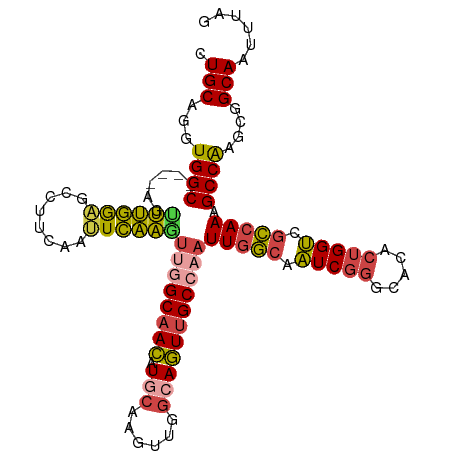
| Location | 630,045 – 630,160 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 86.03 |
| Shannon entropy | 0.27387 |
| G+C content | 0.48766 |
| Mean single sequence MFE | -42.37 |
| Consensus MFE | -30.97 |
| Energy contribution | -34.33 |
| Covariance contribution | 3.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.974208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
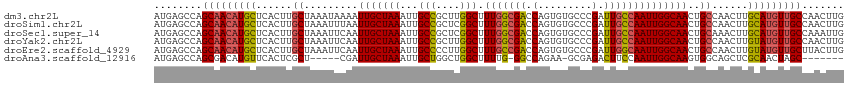
>dm3.chr2L 630045 115 + 23011544 CAAGUUGGCAACAUGCAAGUUGGCAGUUGCCAAUUGGCAAUCGGGCACACUGGUCGCCAAAGCCAAGCGGCAAUUUAGCAAUUUUAUUUAGCAAGUGAGCAUGUUGCUGGCUCAU ..(((..(((((((((.(((((((....)))))))(((.(((((.....))))).)))...(((....)))...........................)))))))))..)))... ( -45.60, z-score = -3.14, R) >droSim1.chr2L 636529 115 + 22036055 CAAGUUGGCAACAUGCAAGUUGGCAGUUGCCAAUUGGCAAUCGGGCACACUGGUCGCCAAAGCCGAGCGGCAAUUUAGCAAUUAAAUUUAGCAAGUGAGCAUGUUGCUGGCUCAU ..(((..(((((((((.(((((((....)))))))(((.(((((.....))))).)))...(((....)))...........................)))))))))..)))... ( -46.10, z-score = -3.24, R) >droSec1.super_14 612231 115 + 2068291 CAAUUUGGCAACAUGCAAGUUUGCAGUUGCCAAUUGGCAAUCGGGCACACUGGUCGCCAAAGCCGAGCGGCAAUUUAGCAAUUGAAUUUAGCAAGUGAGCAUGUUGCUGGCUCAU ....((((((((.((((....))))))))))))(((((.(((((.....))))).)))))((((.(((((((((((((...))))))...((......)).)))))))))))... ( -43.20, z-score = -2.29, R) >droYak2.chr2L 620031 115 + 22324452 CAAGUUGGCAACAUACAAGUUGGCAGUUGCCAAUUGGCAAUCGGGCACACUGGUCGCCAAAGCCAAGCGGCAAUUUAGCAAUUGAAUUUAGCAAGUGAGCAUGUUGCUGGCUCAU ..(((..(((((((.........(((((((...(((((.(((((.....))))).))))).(((....)))......)))))))......((......)))))))))..)))... ( -42.20, z-score = -2.43, R) >droEre2.scaffold_4929 688033 115 + 26641161 CAAGUAAGCAACAUACAAGUUGGCAGUUGCCAAUUGCCAAUCGGGCACACUGGUCGGCAAAGCCAAGGGGCAAUUUAGCAAUUGAAUUUAGCAAGUGAGCAUGUUGCUGGCUCAU ..(((.((((((((.........(((((((...(((((.(((((.....))))).))))).(((....)))......)))))))......((......)))))))))).)))... ( -39.00, z-score = -2.10, R) >droAna3.scaffold_12916 1624541 101 + 16180835 -------GCUAGUUGCGAGCUGCCACUUGCCAAUUGGAAGUCUCGC-UUCUGGCC-CAAAAGCCAGCCAGCAAUUUAGCAAUCG-----AGCGAGUGAACAUGUCGCUGGCUCAU -------(((((((((((((.(..((((.((....))))))..)))-))(((((.-.....)))))...)))).)))))....(-----(((.(((((.....))))).)))).. ( -38.10, z-score = -2.17, R) >consensus CAAGUUGGCAACAUGCAAGUUGGCAGUUGCCAAUUGGCAAUCGGGCACACUGGUCGCCAAAGCCAAGCGGCAAUUUAGCAAUUGAAUUUAGCAAGUGAGCAUGUUGCUGGCUCAU ..((((((((((((((..((((((((((((...(((((.(((((.....))))).))))).(((....)))......)))))))...)))))......))))))))))))))... (-30.97 = -34.33 + 3.36)

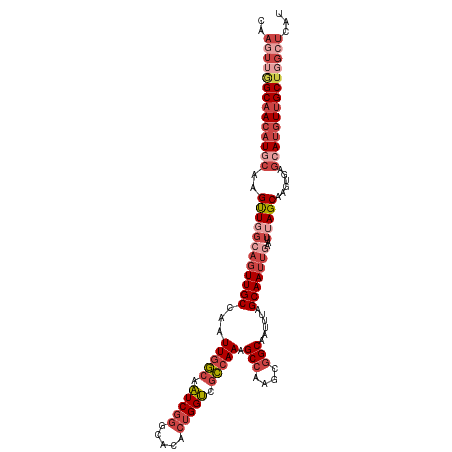
| Location | 630,045 – 630,160 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 86.03 |
| Shannon entropy | 0.27387 |
| G+C content | 0.48766 |
| Mean single sequence MFE | -35.45 |
| Consensus MFE | -26.47 |
| Energy contribution | -26.37 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.763976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
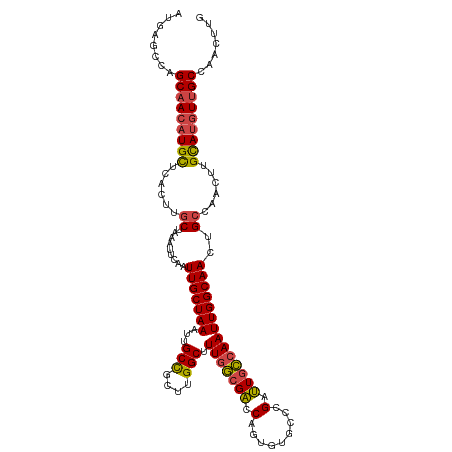
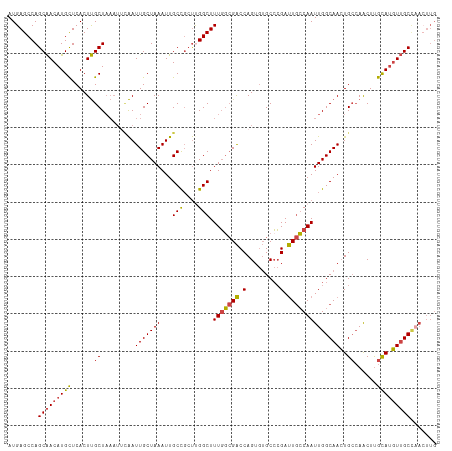
>dm3.chr2L 630045 115 - 23011544 AUGAGCCAGCAACAUGCUCACUUGCUAAAUAAAAUUGCUAAAUUGCCGCUUGGCUUUGGCGACCAGUGUGCCCGAUUGCCAAUUGGCAACUGCCAACUUGCAUGUUGCCAACUUG ..(((...(((((((((......((...........))....((((((.(((((...((((.......)))).....))))).))))))..........)))))))))...))). ( -35.00, z-score = -1.63, R) >droSim1.chr2L 636529 115 - 22036055 AUGAGCCAGCAACAUGCUCACUUGCUAAAUUUAAUUGCUAAAUUGCCGCUCGGCUUUGGCGACCAGUGUGCCCGAUUGCCAAUUGGCAACUGCCAACUUGCAUGUUGCCAACUUG ..(((...(((((((((.................((((((((..(((....)))))))))))..(((..((....(((((....)))))..))..))).)))))))))...))). ( -34.60, z-score = -1.51, R) >droSec1.super_14 612231 115 - 2068291 AUGAGCCAGCAACAUGCUCACUUGCUAAAUUCAAUUGCUAAAUUGCCGCUCGGCUUUGGCGACCAGUGUGCCCGAUUGCCAAUUGGCAACUGCAAACUUGCAUGUUGCCAAAUUG ........(((((((((....((((.........((((((((..(((....))))))))))).............(((((....)))))..))))....)))))))))....... ( -36.30, z-score = -1.77, R) >droYak2.chr2L 620031 115 - 22324452 AUGAGCCAGCAACAUGCUCACUUGCUAAAUUCAAUUGCUAAAUUGCCGCUUGGCUUUGGCGACCAGUGUGCCCGAUUGCCAAUUGGCAACUGCCAACUUGUAUGUUGCCAACUUG ..(((...(((((((((......((...........))....((((((.(((((...((((.......)))).....))))).))))))..........)))))))))...))). ( -33.00, z-score = -1.11, R) >droEre2.scaffold_4929 688033 115 - 26641161 AUGAGCCAGCAACAUGCUCACUUGCUAAAUUCAAUUGCUAAAUUGCCCCUUGGCUUUGCCGACCAGUGUGCCCGAUUGGCAAUUGGCAACUGCCAACUUGUAUGUUGCUUACUUG ..(((..((((((((((..............(((((....)))))....(((((.(((((((......((((.....)))).)))))))..)))))...))))))))))..))). ( -36.30, z-score = -3.00, R) >droAna3.scaffold_12916 1624541 101 - 16180835 AUGAGCCAGCGACAUGUUCACUCGCU-----CGAUUGCUAAAUUGCUGGCUGGCUUUUG-GGCCAGAA-GCGAGACUUCCAAUUGGCAAGUGGCAGCUCGCAACUAGC------- ........((((..(((.((((.((.-----((((((.....(((((..((((((....-)))))).)-))))......)))))))).)))))))..)))).......------- ( -37.50, z-score = -1.40, R) >consensus AUGAGCCAGCAACAUGCUCACUUGCUAAAUUCAAUUGCUAAAUUGCCGCUUGGCUUUGGCGACCAGUGUGCCCGAUUGCCAAUUGGCAACUGCCAACUUGCAUGUUGCCAACUUG ..((((((((((.........)))))..........((......)).....)))))((((((.(.........).)))))).(((((((((((......))).)))))))).... (-26.47 = -26.37 + -0.10)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:06:25 2011