
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 800,283 – 800,341 |
| Length | 58 |
| Max. P | 0.611530 |

| Location | 800,283 – 800,341 |
|---|---|
| Length | 58 |
| Sequences | 10 |
| Columns | 71 |
| Reading direction | reverse |
| Mean pairwise identity | 65.46 |
| Shannon entropy | 0.68363 |
| G+C content | 0.42411 |
| Mean single sequence MFE | -20.43 |
| Consensus MFE | -1.43 |
| Energy contribution | -1.53 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.07 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.611530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
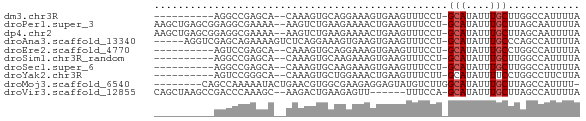
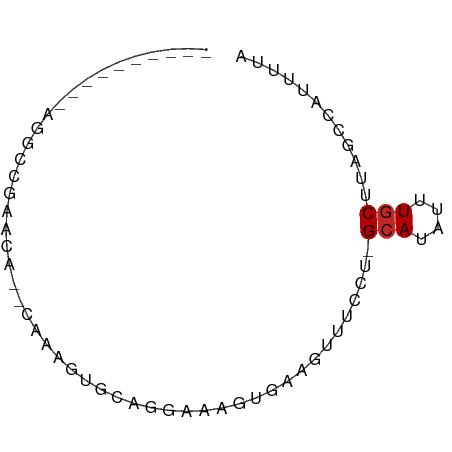
>dm3.chr3R 800283 58 - 27905053 ----------AGGCCGAGCA--CAAAGUGCAGGAAAGUGAAGUUUCCU-GCAUAUUUGCUUGGCCAUUUUA ----------.(((((((((--.((.((((((((((......))))))-)))).)))))))))))...... ( -31.90, z-score = -7.07, R) >droPer1.super_3 2593783 68 + 7375914 AAGCUGAGCGGAGGCGAAAA--AAGUCUGAAGAAAACUGAAGUUUCCU-GCAUAUUUGCUUAGCAAUUUUA ..((((((((((.(((..((--(..((...........))..)))..)-))...))))))))))....... ( -19.10, z-score = -1.91, R) >dp4.chr2 8793553 68 + 30794189 AAGCUGAGCGGAGGCGAAAA--AAGUCUGAAGAAAACUGAAGUUUCCU-GCAUAUUUGCUUAGCAAUUUUA ..((((((((((.(((..((--(..((...........))..)))..)-))...))))))))))....... ( -19.10, z-score = -1.91, R) >droAna3.scaffold_13340 23017531 65 - 23697760 -----AGGUCGAGCAGAAAAGUCUCAGGAAAGUGAAGUGAAGUUUCCU-GCAUAUUUGCCCAGCCAUUUUA -----.(((.(.(((((.......(((((((.(.......).))))))-)....))))).).)))...... ( -16.50, z-score = -1.90, R) >droEre2.scaffold_4770 1095137 58 - 17746568 ----------AGUCCGAGCA--CAAAGUGCAGGAAAGUGAAGUUUCCU-GCAUAUUUGCCUGGCCAUUUUA ----------.(.(((.(((--.((.((((((((((......))))))-)))).))))).))).)...... ( -20.30, z-score = -3.35, R) >droSim1.chr3R_random 180202 58 - 1307089 ----------AGGCCGAGCA--CAAAGUGCAAGAAAGUGAAGUUUCCU-GCAUAUUUGCUUGGCCAUUUUA ----------.(((((((((--.((.(((((.((((......)))).)-)))).)))))))))))...... ( -25.40, z-score = -5.07, R) >droSec1.super_6 944586 58 - 4358794 ----------AGGCCGAGCA--CAAAGUGCAAGAAAGUGAAGUUUCCU-GCAUAUUUGCUUGGCCAUUUUA ----------.(((((((((--.((.(((((.((((......)))).)-)))).)))))))))))...... ( -25.40, z-score = -5.07, R) >droYak2.chr3R 1142520 58 - 28832112 ----------AGUCCGGGCA--CAAAGUGCUGGAAACUGAAGUUUCUU-GCAUAUUUUCCUGGCCUUCUUA ----------((.(((((..--.((.((((.((((((....)))))).-)))).))..))))).))..... ( -18.00, z-score = -2.16, R) >droMoj3.scaffold_6540 10830502 62 + 34148556 --------CAGCCAAAAAUACUGAACGUGGCGAAGAGGAGUAUGUCUUGGCAUAUUUGCUUAGCCAUUUU- --------..................(((((.(((..((((((((....)))))))).))).)))))...- ( -16.80, z-score = -1.90, R) >droVir3.scaffold_12855 2080349 62 + 10161210 CAGCUAAGCCGACCCAAAGC--AAGACUGAAGAGUU------UUUCCA-GCAUAUUUGCUUAGCCAUUUUA ..(((((((...........--((((((....))))------))....-........)))))))....... ( -11.80, z-score = -1.63, R) >consensus __________AGGCCGAACA__CAAAGUGCAGGAAAGUGAAGUUUCCU_GCAUAUUUGCUUAGCCAUUUUA .................................................(((....)))............ ( -1.43 = -1.53 + 0.10)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:50:23 2011