| Sequence ID | dm3.chr3R |
|---|---|
| Location | 773,638 – 773,777 |
| Length | 139 |
| Max. P | 0.633511 |

| Location | 773,638 – 773,777 |
|---|---|
| Length | 139 |
| Sequences | 12 |
| Columns | 168 |
| Reading direction | forward |
| Mean pairwise identity | 88.46 |
| Shannon entropy | 0.23107 |
| G+C content | 0.49565 |
| Mean single sequence MFE | -40.56 |
| Consensus MFE | -30.60 |
| Energy contribution | -30.49 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.536045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
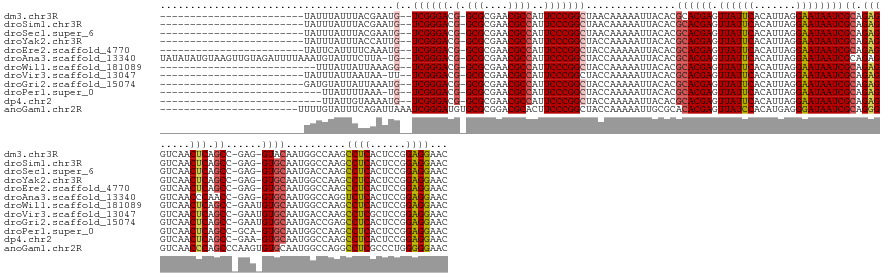
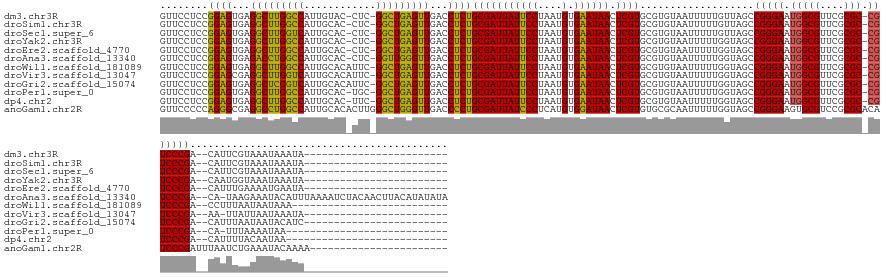
>dm3.chr3R 773638 139 + 27905053 ------------------------UAUUUAUUUACGAAUG--UCGGGACG-GCGCGAACGCCAUUCCCGGCUAACAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGGUCAACUCAGCC-GAG-GUACAAUGGCCAAGCCUCACUCCGGAGGAAC ------------------------..........((...(--((((((.(-(((....))))..)))))))...............((.(((.((((((.......)))))))))..(((.....))).)))-).(-((......)))...((((......))))... ( -37.10, z-score = -1.50, R) >droSim1.chr3R 818967 139 + 27517382 ------------------------UAUUUAUUUACGAAUG--UCGGGACG-GCGCGAACGCCAUUCCCGGCUAACAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGGUCAACUCAGCC-GAG-GUGCAAUGGCCAAGCCUCACUCCGGAGGAAC ------------------------...............(--((((((.(-(((....))))..)))))))...............((((((.((((((.......))))))))((.(((.....))).)).-...-))))..........((((......))))... ( -38.60, z-score = -1.43, R) >droSec1.super_6 903946 139 + 4358794 ------------------------UAUUUAUUUACGAAUG--UCGGGACG-GCGCGAACGCCAUUCCCGGCUAACAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGGUCAACUCAGCC-GAG-GUGCAAUGACCAAGCCUCACUCCGGAGGAAC ------------------------...............(--((((((.(-(((....))))..)))))))...............((((((.((((((.......))))))))((.(((.....))).)).-...-))))..........((((......))))... ( -38.60, z-score = -1.86, R) >droYak2.chr3R 1111984 139 + 28832112 ------------------------UAUUUAUUUACCAUUG--UCGGGACG-GCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGGUCAACUCAGCC-GAG-GUGCAAUGGCCAAGCCUCACUCCGGAGGAAC ------------------------..........((((((--((((((.(-(((....))))..)))))))...............((((((.((((((.......))))))))((.(((.....))).)).-...-))))))))).....((((......))))... ( -42.80, z-score = -2.60, R) >droEre2.scaffold_4770 1070970 139 + 17746568 ------------------------UAUUCAUUUUCAAAUG--UCGGGACG-GCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGGUCAACUCAGCC-GAG-GUGCAAUGGCCAAGCCUCACUCCGGAGGAAC ------------------------...(((((.......(--((((((.(-(((....))))..)))))))...............((((((.((((((.......))))))))((.(((.....))).)).-...-))))))))).....((((......))))... ( -39.70, z-score = -1.65, R) >droAna3.scaffold_13340 22990993 162 + 23697760 UAUAUAUGUAAGUUGUAGAUUUUAAAUGUAUUUCUUA-UG--UCGGGACG-GCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGGUCAACCCAACC-GAG-GUGCAAUGGCCAGGUCUCACUCCGGAGGAAC .......(((((..(((.((.....)).)))..))))-)(--((((((.(-(((....))))..))))))).(((..............(((.((((((.......)))))))))....(((((((((....-).)-))....))))).)))(((......))).... ( -40.10, z-score = -0.04, R) >droWil1.scaffold_181089 6480890 138 + 12369635 --------------------------UUUAUUAUUAAAGG--UCGGGACG-GCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGGUCAACUCAGCC-GAAUGUGCAAUGGCCAAGCCUCACUCCGGAGGAAC --------------------------............((--((((((.(-(((....))))..))))))))..............((((((.((((((.......))))))))((.(((.....))).)).-....))))..........((((......))))... ( -40.00, z-score = -1.95, R) >droVir3.scaffold_13047 11554816 139 + 19223366 ------------------------UAUUUAUUAAUAA-UU--UCGGGACG-GCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGGUCAACUCAGCC-GAAUGUGCAAUGACCAAGCCUCGCUCCGGAGGAAC ------------------------...(((((.....-..--((((((.(-(((....))))..))))))................((((((.((((((.......))))))))((.(((.....))).)).-....))))))))).....((((......))))... ( -35.70, z-score = -1.23, R) >droGri2.scaffold_15074 1426149 140 - 7742996 ------------------------GAUGUAUUAUUAAAUG--UCGGGACG-GCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGGUCAACUCAGCC-GAAUGUGCAAUGACCGAGCCUCACUCCGGAGGAAC ------------------------..((((.........(--((((((.(-(((....))))..)))))))..........))))....((.((((((((((((..((....))((.(((.....))).)).-.)))))).))))))))..((((......))))... ( -41.51, z-score = -2.09, R) >droPer1.super_0 4291594 135 - 11822988 ---------------------------UUAUUUUAAA-UG--UCGGGACG-GCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGGUCAACUCAGCC-GCA-GUGCAAUGGCCAAGCCUCACUCCGGAGGAAC ---------------------------..........-.(--((((((.(-(((....))))..)))))))..((..............(((.((((((.......)))))))))..(((.....)))..((-(.(-(((....(((...))).)))).))).))... ( -39.00, z-score = -1.83, R) >dp4.chr2 17866257 136 - 30794189 ---------------------------UUAUUGUAAAAUG--UCGGGACG-GCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGGUCAACUCAGCC-GAA-GUGCAAUGGCCAAGCCUCACUCCGGAGGAAC ---------------------------............(--((((((.(-(((....))))..)))))))..((..............(((.((((((.......)))))))))..(((.....)))..((-(.(-(((....(((...))).)))).))).))... ( -39.00, z-score = -1.56, R) >anoGam1.chr2R 55419100 145 - 62725911 -----------------------UUUUGUAUUUCAGAUUAAAUCGGGAUGUGCGCGGACGCACUUCCCGGCUACCAAAAAUUGCGCACACGAGUUAUCCACAUGAGGGAUAAUCGCAGGGGUCAACCCAGCCCAAGUGUGCAAUGGCCAGGCCUCGCCCUGGGGGAAC -----------------------........(((........((((((.(((((....))))).))))))...((....(((((((((..((.((((((.......))))))))((.(((.....))).))....)))))))))..(((((......)))))))))). ( -54.60, z-score = -1.99, R) >consensus ________________________UAUUUAUUUACAAAUG__UCGGGACG_GCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGGUCAACUCAGCC_GAG_GUGCAAUGGCCAAGCCUCACUCCGGAGGAAC ..........................................((((((...(((....)))...))))))................((((((.((((((.......))))))))((.(((.....))).))......))))..........((((......))))... (-30.60 = -30.49 + -0.11)
| Location | 773,638 – 773,777 |
|---|---|
| Length | 139 |
| Sequences | 12 |
| Columns | 168 |
| Reading direction | reverse |
| Mean pairwise identity | 88.46 |
| Shannon entropy | 0.23107 |
| G+C content | 0.49565 |
| Mean single sequence MFE | -44.75 |
| Consensus MFE | -36.82 |
| Energy contribution | -35.75 |
| Covariance contribution | -1.07 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.633511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
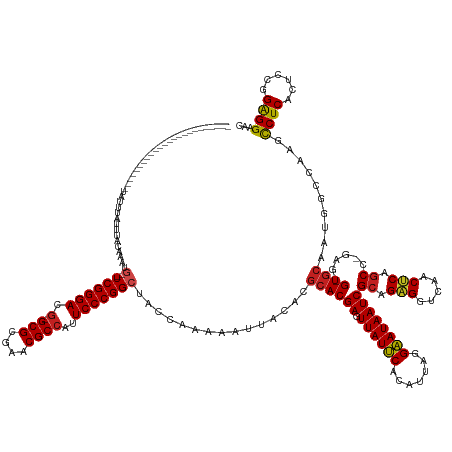
>dm3.chr3R 773638 139 - 27905053 GUUCCUCCGGAGUGAGGCUUGGCCAUUGUAC-CUC-GGCUGAGUUGACCUCUGCGAUUAUUCCUAAUGUGAAUAACUCGUGCGUGUAAUUUUUGUUAGCCGGGAAUGGCGUUCGCGC-CGUCCCGA--CAUUCGUAAAUAAAUA------------------------ .......(((((...((((..(((.......-...-)))..))))...)))))((((((((((....).)))))).)))((((..((((....))))(.(((((.(((((....)))-))))))).--)...))))........------------------------ ( -42.20, z-score = -1.37, R) >droSim1.chr3R 818967 139 - 27517382 GUUCCUCCGGAGUGAGGCUUGGCCAUUGCAC-CUC-GGCUGAGUUGACCUCUGCGAUUAUUCCUAAUGUGAAUAACUCGUGCGUGUAAUUUUUGUUAGCCGGGAAUGGCGUUCGCGC-CGUCCCGA--CAUUCGUAAAUAAAUA------------------------ .........(((((.((((.(((.(((((((-(.(-(((.(((.....))).))..(((((((....).))))))..)).).)))))))....)))))))((((.(((((....)))-))))))..--)))))...........------------------------ ( -44.00, z-score = -1.63, R) >droSec1.super_6 903946 139 - 4358794 GUUCCUCCGGAGUGAGGCUUGGUCAUUGCAC-CUC-GGCUGAGUUGACCUCUGCGAUUAUUCCUAAUGUGAAUAACUCGUGCGUGUAAUUUUUGUUAGCCGGGAAUGGCGUUCGCGC-CGUCCCGA--CAUUCGUAAAUAAAUA------------------------ .........(((((.((((.....(((((((-(.(-(((.(((.....))).))..(((((((....).))))))..)).).))))))).......))))((((.(((((....)))-))))))..--)))))...........------------------------ ( -43.40, z-score = -1.60, R) >droYak2.chr3R 1111984 139 - 28832112 GUUCCUCCGGAGUGAGGCUUGGCCAUUGCAC-CUC-GGCUGAGUUGACCUCUGCGAUUAUUCCUAAUGUGAAUAACUCGUGCGUGUAAUUUUUGGUAGCCGGGAAUGGCGUUCGCGC-CGUCCCGA--CAAUGGUAAAUAAAUA------------------------ ...((((......))))....((((((((..-...-(((((((.....))).(((((((((((....).)))))).))))................))))((((.(((((....)))-))))))).--))))))).........------------------------ ( -44.20, z-score = -1.16, R) >droEre2.scaffold_4770 1070970 139 - 17746568 GUUCCUCCGGAGUGAGGCUUGGCCAUUGCAC-CUC-GGCUGAGUUGACCUCUGCGAUUAUUCCUAAUGUGAAUAACUCGUGCGUGUAAUUUUUGGUAGCCGGGAAUGGCGUUCGCGC-CGUCCCGA--CAUUUGAAAAUGAAUA------------------------ ((((....((((...((((..(((.......-...-)))..))))...))))(((((((((((....).)))))).)))).......(((((..(..(.(((((.(((((....)))-))))))).--)..)..))))))))).------------------------ ( -45.00, z-score = -1.56, R) >droAna3.scaffold_13340 22990993 162 - 23697760 GUUCCUCCGGAGUGAGACCUGGCCAUUGCAC-CUC-GGUUGGGUUGACCUCUGCGAUUAUUCCUAAUGUGAAUAACUCGUGCGUGUAAUUUUUGGUAGCCGGGAAUGGCGUUCGCGC-CGUCCCGA--CA-UAAGAAAUACAUUUAAAAUCUACAACUUACAUAUAUA ........((((...((((..(((.......-...-)))..))))...))))(((((((((((....).)))))).))))..(((((.((((((...(.(((((.(((((....)))-))))))).--).-)))))).)))))......................... ( -47.40, z-score = -1.54, R) >droWil1.scaffold_181089 6480890 138 - 12369635 GUUCCUCCGGAGUGAGGCUUGGCCAUUGCACAUUC-GGCUGAGUUGACCUCUGCGAUUAUUCCUAAUGUGAAUAACUCGUGCGUGUAAUUUUUGGUAGCCGGGAAUGGCGUUCGCGC-CGUCCCGA--CCUUUAAUAAUAAA-------------------------- (((((...((((...((((..(((...........-)))..))))...))))(((((((((((....).)))))).)))).............)).)))(((((.(((((....)))-))))))).--..............-------------------------- ( -43.10, z-score = -1.37, R) >droVir3.scaffold_13047 11554816 139 - 19223366 GUUCCUCCGGAGCGAGGCUUGGUCAUUGCACAUUC-GGCUGAGUUGACCUCUGCGAUUAUUCCUAAUGUGAAUAACUCGUGCGUGUAAUUUUUGGUAGCCGGGAAUGGCGUUCGCGC-CGUCCCGA--AA-UUAUUAAUAAAUA------------------------ (((((...)))))..((((..(..(((((((..((-(((...))))).....(((((((((((....).)))))).))))..)))))))..)..)..)))((((.(((((....)))-))))))..--..-.............------------------------ ( -43.00, z-score = -1.30, R) >droGri2.scaffold_15074 1426149 140 + 7742996 GUUCCUCCGGAGUGAGGCUCGGUCAUUGCACAUUC-GGCUGAGUUGACCUCUGCGAUUAUUCCUAAUGUGAAUAACUCGUGCGUGUAAUUUUUGGUAGCCGGGAAUGGCGUUCGCGC-CGUCCCGA--CAUUUAAUAAUACAUC------------------------ ........((((...(((((((((...........-)))))))))...))))(((((((((((....).)))))).))))..((((.(((....((...(((((.(((((....)))-))))))))--)....))).))))...------------------------ ( -43.20, z-score = -1.12, R) >droPer1.super_0 4291594 135 + 11822988 GUUCCUCCGGAGUGAGGCUUGGCCAUUGCAC-UGC-GGCUGAGUUGACCUCUGCGAUUAUUCCUAAUGUGAAUAACUCGUGCGUGUAAUUUUUGGUAGCCGGGAAUGGCGUUCGCGC-CGUCCCGA--CA-UUUAAAAUAA--------------------------- .........(((((.((((..((((((((((-.((-(((.(((.....))).))(((((((((....).)))))).)).)))))))))....))))))))((((.(((((....)))-))))))..--))-))).......--------------------------- ( -45.70, z-score = -1.92, R) >dp4.chr2 17866257 136 + 30794189 GUUCCUCCGGAGUGAGGCUUGGCCAUUGCAC-UUC-GGCUGAGUUGACCUCUGCGAUUAUUCCUAAUGUGAAUAACUCGUGCGUGUAAUUUUUGGUAGCCGGGAAUGGCGUUCGCGC-CGUCCCGA--CAUUUUACAAUAA--------------------------- ........((((((.((((..((((((((((-.((-(((...))))).....(((((((((((....).)))))).))))..))))))....))))))))((((.(((((....)))-))))))..--)))))).......--------------------------- ( -43.70, z-score = -1.50, R) >anoGam1.chr2R 55419100 145 + 62725911 GUUCCCCCAGGGCGAGGCCUGGCCAUUGCACACUUGGGCUGGGUUGACCCCUGCGAUUAUCCCUCAUGUGGAUAACUCGUGUGCGCAAUUUUUGGUAGCCGGGAAGUGCGUCCGCGCACAUCCCGAUUUAAUCUGAAAUACAAAA----------------------- .......((((((...(((.......(((((((....((.(((.....))).))(((((((((....).)))))).)))))))))........))).)))((((.(((((....))))).))))........)))..........----------------------- ( -52.16, z-score = -1.28, R) >consensus GUUCCUCCGGAGUGAGGCUUGGCCAUUGCAC_CUC_GGCUGAGUUGACCUCUGCGAUUAUUCCUAAUGUGAAUAACUCGUGCGUGUAAUUUUUGGUAGCCGGGAAUGGCGUUCGCGC_CGUCCCGA__CAUUUGAAAAUAAAUA________________________ ........((((...(((((((((............)))))))))...))))(((((((((((....).)))))).))))...................(((((...(((....)))...)))))........................................... (-36.82 = -35.75 + -1.07)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:50:18 2011