
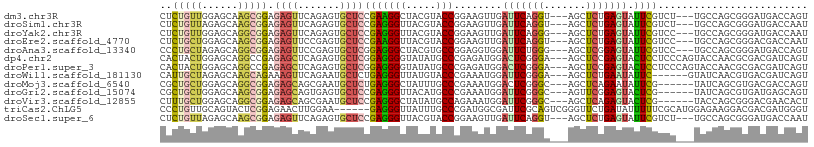
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 763,006 – 763,108 |
| Length | 102 |
| Max. P | 0.573914 |

| Location | 763,006 – 763,108 |
|---|---|
| Length | 102 |
| Sequences | 13 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 74.40 |
| Shannon entropy | 0.54951 |
| G+C content | 0.55574 |
| Mean single sequence MFE | -37.85 |
| Consensus MFE | -14.56 |
| Energy contribution | -14.15 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.78 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.573914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
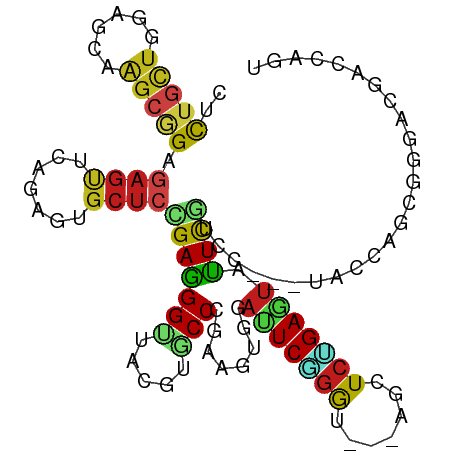
>dm3.chr3R 763006 102 + 27905053 CUCUGUUGGAGCAAGCGGAGAGUUCAGAGUGCUCCGAAGGCUACGUACCGGAAGUUGAUUCAGGU---AGCUCUGAGUAUUCGUCU---UGCCAGCGGGAUGACCAGU .(((((((..(((((((((...(((((((((((.....)))....((((.(((.....))).)))---)))))))))..)))).))---))))))))))......... ( -35.60, z-score = -1.11, R) >droSim1.chr3R 804029 102 + 27517382 CUCUGUUAGAGCAAGCGGAGAGUUCAGAGUGCUCCGAGGGUUACGUACCGGAAGUUGAUUCAGGU---AGCUCUGAGUAUUCGUCU---UGCCAGCGGGAUGACCAAU .((((((.(.(((((((((...((((((((...((...(((.....))).(((.....))).)).---.))))))))..)))).))---))))))))))......... ( -33.50, z-score = -1.15, R) >droYak2.chr3R 1084516 102 + 28832112 CUCUGUUGGAGCAGGCGGAGAGUUCAGAGUGCUCCGAGGGUUACGUACCGGAAGUUGAUUCAGGG---AGCUCUGAGUAUUCGUCC---UGCCAGCGGGAUGACCAAU .(((((((..(((((((((...((((((..(((((...(((.....))).(((.....)))..))---)))))))))..)))).))---))))))))))......... ( -38.40, z-score = -1.53, R) >droEre2.scaffold_4770 1061267 102 + 17746568 CUCUGCUGGAGCAAGCGGAGAGUUCCGAGUGCUCCGAAGGUUACGUACCGGAAGUUGAUUCAGGU---AGCUCUGAGUAUUCGUCC---UGCCAGCGGGACGACCAAU ......(((((((..(((......)))..)))))))..(((.....)))((......(((((((.---...)))))))..((((((---((....))))))))))... ( -41.50, z-score = -2.64, R) >droAna3.scaffold_13340 22974648 102 + 23697760 CCCUGCUAGAGCAGGCGGAGAGUUCCGAGUGCUCGGAGGGCUACGUGCCGGAGGUGGAUUCUGGG---AGCUCGGAGUAUUCGUCC---UGCCAGCGGGAUGACCAGU .((((((.(.(((((((((...((((((((.(((((((..((((.(.....).)))).)))))))---.))))))))..)))).))---))))))))))......... ( -49.50, z-score = -2.57, R) >dp4.chr2 8764391 105 - 30794189 CACUACUGGAGCAGGCCGAGAGCUCAGAGUGCUCGGAGGGGUAUAUGCCCGAGAUGGACUCGGGA---AGCUCCGAGUACUCCUCCCAGUACCAACGCGACGAUCAGU ...(((((((((.........)))).((((((((((((((((....))))(((.....)))....---..))))))))))))....)))))................. ( -40.00, z-score = -1.12, R) >droPer1.super_3 2564685 105 - 7375914 CACUACUGGAGCAGGCCGAGAGCUCAGAGUGCUCGGAGGGGUAUAUGCCCGAGAUGGACUCGGGA---AGCUCCGAGUACUCCUCCCAGUACCAACGCGACGAUCAGU ...(((((((((.........)))).((((((((((((((((....))))(((.....)))....---..))))))))))))....)))))................. ( -40.00, z-score = -1.12, R) >droWil1.scaffold_181130 8222695 99 - 16660200 CAUUGCUAGAGCAAGCAGAAAGUUCAGAAUGCUCUGAGGGUUAUGUACCCGAAAUGGAUUCGGGA---AGCUCUGAAUAUUC------GUAUCAACGUGACGAUCAGU ......((((((..(((.((..((((((....))))))..)).))).((((((.....)))))).---.)))))).....((------((.........))))..... ( -26.80, z-score = -1.23, R) >droMoj3.scaffold_6540 23735934 99 + 34148556 CGCUGCUGGAGCAGGCGGAGAGCAGCGAAUGCUCUGAGGGCUAUUUGCCCGAAAUGGACUCGGGC---AGCUCAGAAUAUUCG------UAUCAGCGUGACGACCAGU ....(((((.(((.((...((...(((((((.((((((......((((((((.......))))))---)))))))).))))))------).)).)).)).)..))))) ( -39.30, z-score = -1.77, R) >droGri2.scaffold_15074 5118624 99 + 7742996 CGCUGCUGGAGCAAGCGGAGAGCAGUGAGUGCUCCGAGGGUUACAUGCCCGAAAUGGAUUCGGGC---AGUUCGGAGUACUCG------UAUCAGCGUGAUGAGCAGU (((.(((((.((..((.....))...(((((((((((((....).((((((((.....)))))))---).)))))))))))))------).))))))))......... ( -45.40, z-score = -4.21, R) >droVir3.scaffold_12855 8536826 99 + 10161210 CUUUGCUGGAGCAGGCGGAGAGCAGCGAAUGCUCCGAGGGCUAUAUGCCAGAAAUGGAUUCGGGC---AGCUCAGAGUACUCG------UACCAGCGGGACGAACACU .((((((((.....((.....)).((((.(((((.(((.(((.....(((....))).....)))---..))).))))).)))------).))))))))......... ( -37.20, z-score = -1.95, R) >triCas2.ChLG5 13793123 102 - 18847211 CCCUGUUGCAGUACUCGGAGAACUUGGAA------GAGGGUUAUUUGCCCGAUGGCGAUUCGCAGUCGGGUUCUGAUAUUUUUCGCAUGGAGAAGGACGACGAUGGGU .(((((((..((..(((((.(((((....------..)))))....(((((((.(((...))).))))))))))))..((((((.....)))))).))..))))))). ( -31.30, z-score = -0.48, R) >droSec1.super_6 862462 102 + 4358794 CUCUGUUAGAGCAAGCGGAGAGUUCAGAGUGCUCCGAGGGUUACGUACCGGAAGUUGAUUCAGGU---AGCUCUGAGUAUUCGUCU---UGCCAGCGGGAUGACCAAU .((((((.(.(((((((((...((((((((...((...(((.....))).(((.....))).)).---.))))))))..)))).))---))))))))))......... ( -33.50, z-score = -1.15, R) >consensus CUCUGCUGGAGCAAGCGGAGAGUUCAGAGUGCUCCGAGGGUUACGUGCCCGAAGUGGAUUCGGGU___AGCUCUGAGUAUUCGUCC___UACCAGCGGGACGACCAGU ..(((((......))))).((((.......))))(((((((.....)))........(((((((.......))))))).))))......................... (-14.56 = -14.15 + -0.41)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:50:15 2011