| Sequence ID | dm3.chr3R |
|---|---|
| Location | 680,726 – 680,818 |
| Length | 92 |
| Max. P | 0.500000 |

| Location | 680,726 – 680,818 |
|---|---|
| Length | 92 |
| Sequences | 10 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 61.32 |
| Shannon entropy | 0.76327 |
| G+C content | 0.40204 |
| Mean single sequence MFE | -25.11 |
| Consensus MFE | -2.72 |
| Energy contribution | -3.40 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.11 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
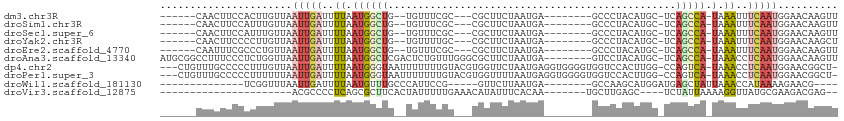
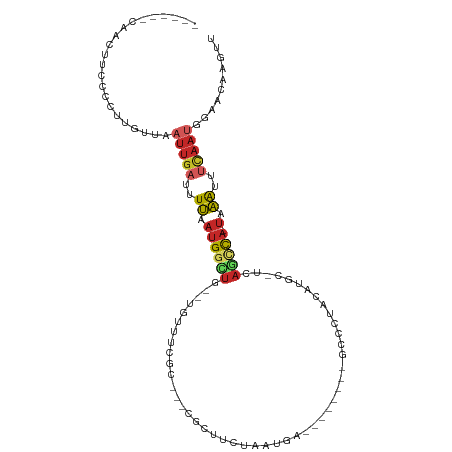
>dm3.chr3R 680726 92 - 27905053 ------CAACUUCCACUUGUUAAUUGAUUUUAAUGGCUG--UGUUUCGC---CGCUUCUAAUGA--------GCCCUACAUGC-UCAGCCA-UAAAUUUCAAUGGAACAAGUU ------........(((((((.(((((..((.(((((((--.((.....---.((((.....))--------)).......))-.))))))-).))..)))))..))))))). ( -28.32, z-score = -4.74, R) >droSim1.chr3R 726899 92 - 27517382 ------CAACUUCCAUUUGUUAAUUGAUUUUAAUGGCUG--UGUUUCGC---CGCUUCUAAUGA--------GCCCUACAUGC-UCAGCCA-UAAAUUUCAAUGGAACAAGUU ------........(((((((.(((((..((.(((((((--.((.....---.((((.....))--------)).......))-.))))))-).))..)))))..))))))). ( -26.02, z-score = -3.88, R) >droSec1.super_6 784369 92 - 4358794 ------CAACUUCCAUUUGUUAAUUGAUUUUAAUGGCUG--UGUUUCGC---CGCUUCUAAUGA--------GCCCUACAUGC-UCAGCCA-UAAAUUUCAAUGGAACAAGUU ------........(((((((.(((((..((.(((((((--.((.....---.((((.....))--------)).......))-.))))))-).))..)))))..))))))). ( -26.02, z-score = -3.88, R) >droYak2.chr3R 1004001 92 - 28832112 ------CAACUUCCCCUUGUUAAUUGAUUUUAAUGGCUG--UGUUUUGC---CGCUUCUAAUGA--------GCCCUACAUGC-UCAGCCA-UAAAUUUCAAUGGAACAAGCU ------.........((((((.(((((..((.(((((((--.((.....---.((((.....))--------)).......))-.))))))-).))..)))))..)))))).. ( -26.92, z-score = -4.16, R) >droEre2.scaffold_4770 986265 92 - 17746568 ------CAAUUUCGCCCUGUUAAUUGAUUUUAAUGGCUG--UGUUUCGC---CGCUUCUAAUGA--------GCCCUACAUGC-UCAGCCA-UAAAUUUCAAUGGAACAAGUU ------...........((((.(((((..((.(((((((--.((.....---.((((.....))--------)).......))-.))))))-).))..)))))..)))).... ( -23.62, z-score = -2.68, R) >droAna3.scaffold_13340 798526 103 + 23697760 AUGCGGCCUUUCCCUCUGGUUAAUUGAUUUUAAUGGCUCGACUCUGUUUGGGCGCUUCUAAUGA--------GUCCUACAUGC-UCAGCCA-UAAACCUCAAUGGAACAAGUU ....((......))....(((.(((((..((.((((((.((.(.(((..((((.(.......).--------)))).))).).-)))))))-).))..)))))..)))..... ( -21.60, z-score = 0.36, R) >dp4.chr2 22112770 107 - 30794189 ---CUGUUUGCCCCCUUUGUUAAUUGAUUUUAAUGGGUAAUUUUUUUGUACGUGGUUCUAAUGAGGUGGGGUGGUCCACUUGG-CCAGUCA-UAAACCUCAAUGGAACGGCU- ---....((((((.....(((((......)))))))))))...........((.((((((.((((((.((.(((((.....))-))).)).-...)))))).)))))).)).- ( -35.70, z-score = -2.59, R) >droPer1.super_3 4893173 107 - 7375914 ---CUGUUUGCCCCCUUUUUUAAUUGAUUUUAAUGGGUAAUUUUUUUGUACGUGGUUUUAAUGAGGUGGGGUGGUCCACUUGG-CCAGUCA-UAAACCUCAAUGGAACGGCU- ---....((((((......((((......)))).))))))...........((.((((((.((((((.((.(((((.....))-))).)).-...)))))).)))))).)).- ( -31.40, z-score = -1.46, R) >droWil1.scaffold_181130 8516298 82 - 16660200 --------------UCGGUUUAAUUGAUUUUAAUGUUUGCCCAUUCCG-----GUUCUUAAUGA--------GCCAAGCAUGGAUGAGCUAUUAAACCAUAAAAGAACG---- --------------..((((((((..........((((..((((.(.(-----((((.....))--------)))..).))))..)))).))))))))...........---- ( -20.10, z-score = -2.45, R) >droVir3.scaffold_12875 2158843 78 - 20611582 ----------------------ACGCCCCUCAGCGCUUCACUAUUUUUGAAACAUAUUUCACAA-------UGCUUGAGC----UCUAUUAAAAGGUUAUGCGAAGACGAG-- ----------------------.(((......)))((((........(((((....)))))...-------.((.(((.(----(........)).))).)))))).....-- ( -11.40, z-score = 0.23, R) >consensus ______CAACUUCCCCUUGUUAAUUGAUUUUAAUGGCUG__UGUUUCGC___CGCUUCUAAUGA________GCCCUACAUGC_UCAGCCA_UAAAUUUCAAUGGAACAAGUU ......................(((((.((((.(((((................................................))))).))))..))))).......... ( -2.72 = -3.40 + 0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:50:08 2011