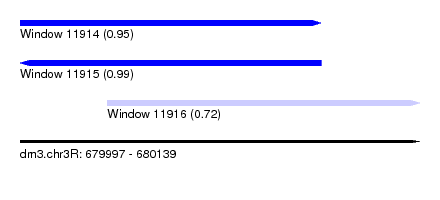
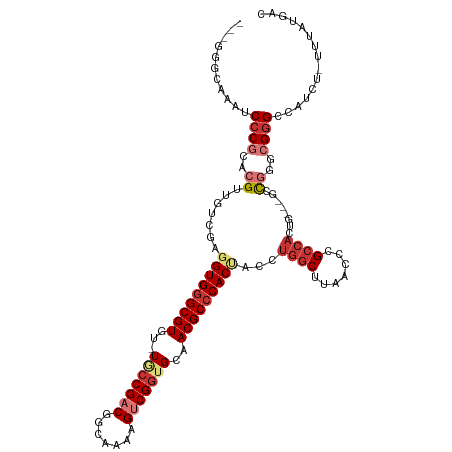
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 679,997 – 680,139 |
| Length | 142 |
| Max. P | 0.992285 |

| Location | 679,997 – 680,104 |
|---|---|
| Length | 107 |
| Sequences | 8 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 82.08 |
| Shannon entropy | 0.33626 |
| G+C content | 0.60291 |
| Mean single sequence MFE | -48.00 |
| Consensus MFE | -28.67 |
| Energy contribution | -29.07 |
| Covariance contribution | 0.41 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.953557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
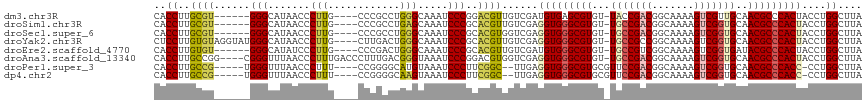
>dm3.chr3R 679997 107 + 27905053 CACCUUGCGU------GGGCAUAACCCUUG----CCCGCCUGGGCAAAUCCCGGACGUUGUCGAUGUGAGCGUGU-UACCGACGGCAAAAGUCGUUGCAACGCCCACUACCUGGCUUA ........((------(((((.......))----)))))(((((.....))))).....((((..(((.((((..-...(((((((....)))))))..)))).)))....))))... ( -38.20, z-score = -1.08, R) >droSim1.chr3R 726140 107 + 27517382 CACCUUGCGU------GGGCAUAACCCUUG----CCCGCCUGAGCAAAUCCCGCACGUUGUCGAGGUGGGCGUGU-UGCCGACGGCAAAAGUCGGUGCAACGCCCACUACCUGGCUUA ........((------(((((.......))----))))).(((((..........((....)).(((((((((..-.((((((.......))))))...))))))))).....))))) ( -45.00, z-score = -1.84, R) >droSec1.super_6 783610 107 + 4358794 CACCUUGCGU------GGGCAUAACCCUUG----CCCGCCUGGGCAAAUCCCGCACGUGGUCGAGGUGGGCGUGU-UGCCGACGGCAAAAGUCGGUGCAACGCCCACUACCUGGCUUA (((..(((((------(((((.......))----)))))..(((.....)))))).)))((((.(((((((((..-.((((((.......))))))...)))))))))...))))... ( -51.30, z-score = -2.40, R) >droYak2.chr3R 1003245 113 + 28832112 CUCCUUGUGUAGGUAUGGGCAUAACCCUUG----CUUGACUGGGCAAAUCCCGCACGUUGUCGAGGUGGGCGUGU-UGCCGCCGGCAAAAGUCGGUGCAACGCCCACUACCUGGCUUA ......(((((((((.(((.....))).))----)))....(((.....)))))))...(((.(((((((.((((-(((.((((((....))))))))))))).).)))))))))... ( -46.60, z-score = -1.64, R) >droEre2.scaffold_4770 985125 107 + 17746568 CACCUUGUGU------GGGCAUAUCCCUUG----CCCGACUGGGCAAAUCCCGCACGUUGUCGAUGUGGGCGUGU-UGCCGUCGGCAAAAGUCGGUGAUACGCCCACUACCUGGCUUA ......((((------(((.((.....(((----(((....)))))))))))))))...((((..((((((((((-..(((.(.......).)))..))))))))))....))))... ( -51.20, z-score = -4.15, R) >droAna3.scaffold_13340 22896975 113 + 23697760 CACCUUGCCGG----CGGGUUUAACCCUUUGACCCUUUGACGGGUAAAUCCCGGACGUGGUCGAGGUGGGCGUGU-UGCCGACGGCAAAAGUCGGUGCAACGCCCACUACCUGGCUUA ......(((((----.(((.....))).((((((......((((.....)))).....))))))(((((((((..-.((((((.......))))))...)))))))))..)))))... ( -53.80, z-score = -3.24, R) >droPer1.super_3 2480348 106 - 7375914 CACCUUGCCG-----UGGGUUUAACCCUUU----CCGGGGCAUGUAAAUCCCUUCGGC--UUGAGGUGGGCGUGCGUUCCGACGGCAAAAGUCGGUGCAACGCCCACC-CCUGGCUUA ..((..((((-----.(((((((.((((..----..))))....)))))))...))))--....(((((((((..((.(((((.......))))).)).)))))))))-...)).... ( -48.50, z-score = -2.81, R) >dp4.chr2 8679005 106 - 30794189 CACCUUGCCG-----UGGGUUUAACCCUUU----CCGGGGCAAGUAAAUCCCUUCGGC--UUGAGGUGGGCGUGCGUUCCGACGGCAAAAGUCGGUGCAACGCCCACC-CCUGGCUUA ...((((((.-----((((..........)----))).))))))...........(((--(.(.(((((((((..((.(((((.......))))).)).)))))))))-.).)))).. ( -49.40, z-score = -3.03, R) >consensus CACCUUGCGU______GGGCAUAACCCUUG____CCCGCCUGGGCAAAUCCCGCACGUUGUCGAGGUGGGCGUGU_UGCCGACGGCAAAAGUCGGUGCAACGCCCACUACCUGGCUUA ................(((.....)))..............((((..........((....)).(((((((((...(((((((.......)))))))..))))))))).....)))). (-28.67 = -29.07 + 0.41)
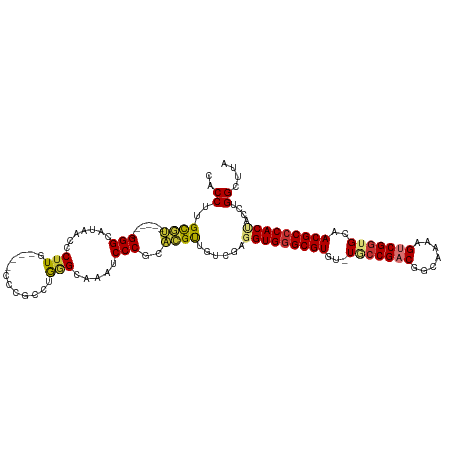
| Location | 679,997 – 680,104 |
|---|---|
| Length | 107 |
| Sequences | 8 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 82.08 |
| Shannon entropy | 0.33626 |
| G+C content | 0.60291 |
| Mean single sequence MFE | -48.22 |
| Consensus MFE | -30.89 |
| Energy contribution | -31.00 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.992285 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 679997 107 - 27905053 UAAGCCAGGUAGUGGGCGUUGCAACGACUUUUGCCGUCGGUA-ACACGCUCACAUCGACAACGUCCGGGAUUUGCCCAGGCGGG----CAAGGGUUAUGCCC------ACGCAAGGUG ...(((..((.((((((((......((((((((((((((((.-..........))))))..((((.(((.....))).))))))----))))))))))))))------))))..))). ( -47.00, z-score = -3.14, R) >droSim1.chr3R 726140 107 - 27517382 UAAGCCAGGUAGUGGGCGUUGCACCGACUUUUGCCGUCGGCA-ACACGCCCACCUCGACAACGUGCGGGAUUUGCUCAGGCGGG----CAAGGGUUAUGCCC------ACGCAAGGUG ...(((..(..((((((((....(((((.......)))))..-..))))))))..).......((((....((((((....)))----)))(((.....)))------.)))).))). ( -44.30, z-score = -1.26, R) >droSec1.super_6 783610 107 - 4358794 UAAGCCAGGUAGUGGGCGUUGCACCGACUUUUGCCGUCGGCA-ACACGCCCACCUCGACCACGUGCGGGAUUUGCCCAGGCGGG----CAAGGGUUAUGCCC------ACGCAAGGUG ...(((.(((.((((((((....(((((.......)))))..-..))))))))....)))...((((....((((((....)))----)))(((.....)))------.)))).))). ( -49.90, z-score = -2.28, R) >droYak2.chr3R 1003245 113 - 28832112 UAAGCCAGGUAGUGGGCGUUGCACCGACUUUUGCCGGCGGCA-ACACGCCCACCUCGACAACGUGCGGGAUUUGCCCAGUCAAG----CAAGGGUUAUGCCCAUACCUACACAAGGAG ....((..(..(((((((((((.(((.(....).)))..)))-..))))))))..)......(((..((..((((........)----)))(((.....)))...))..)))..)).. ( -40.10, z-score = -1.09, R) >droEre2.scaffold_4770 985125 107 - 17746568 UAAGCCAGGUAGUGGGCGUAUCACCGACUUUUGCCGACGGCA-ACACGCCCACAUCGACAACGUGCGGGAUUUGCCCAGUCGGG----CAAGGGAUAUGCCC------ACACAAGGUG ...(((..((.(((((((((((.((.....((((((((((((-(....(((.((.((....)))).)))..)))))..))).))----))))))))))))))------))))..))). ( -49.30, z-score = -4.34, R) >droAna3.scaffold_13340 22896975 113 - 23697760 UAAGCCAGGUAGUGGGCGUUGCACCGACUUUUGCCGUCGGCA-ACACGCCCACCUCGACCACGUCCGGGAUUUACCCGUCAAAGGGUCAAAGGGUUAAACCCG----CCGGCAAGGUG ...(((.(((.((((((((....(((((.......)))))..-..))))))))...((((.....((((.....))))......))))...(((.....))))----)))))...... ( -48.40, z-score = -2.77, R) >droPer1.super_3 2480348 106 + 7375914 UAAGCCAGG-GGUGGGCGUUGCACCGACUUUUGCCGUCGGAACGCACGCCCACCUCAA--GCCGAAGGGAUUUACAUGCCCCGG----AAAGGGUUAAACCCA-----CGGCAAGGUG ...(((.((-(((((((((.((.(((((.......)))))...)))))))))))))..--((((..(((.((((...((((...----...))))))))))).-----))))..))). ( -53.40, z-score = -4.47, R) >dp4.chr2 8679005 106 + 30794189 UAAGCCAGG-GGUGGGCGUUGCACCGACUUUUGCCGUCGGAACGCACGCCCACCUCAA--GCCGAAGGGAUUUACUUGCCCCGG----AAAGGGUUAAACCCA-----CGGCAAGGUG ...(((.((-(((((((((.((.(((((.......)))))...)))))))))))))..--((((..(((.((((...((((...----...))))))))))).-----))))..))). ( -53.40, z-score = -4.31, R) >consensus UAAGCCAGGUAGUGGGCGUUGCACCGACUUUUGCCGUCGGCA_ACACGCCCACCUCGACAACGUGCGGGAUUUGCCCAGCCGGG____CAAGGGUUAUGCCC______ACGCAAGGUG ...(((.....((((((((....(((((.......))))).....)))))))).............(((....((((..............))))....)))............))). (-30.89 = -31.00 + 0.11)
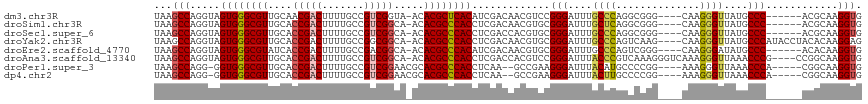
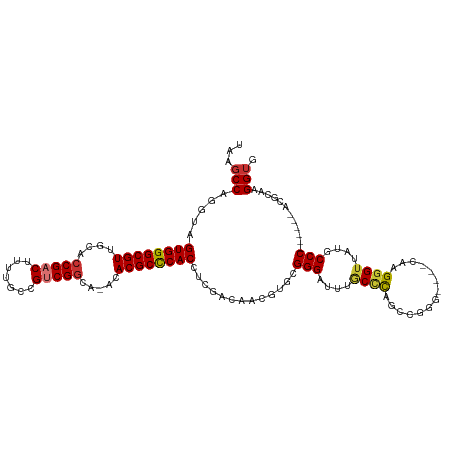
| Location | 680,028 – 680,139 |
|---|---|
| Length | 111 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.94 |
| Shannon entropy | 0.33555 |
| G+C content | 0.60873 |
| Mean single sequence MFE | -45.48 |
| Consensus MFE | -30.21 |
| Energy contribution | -31.90 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.716820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 680028 111 + 27905053 ---GGGCAAAUCCCGGACGUUGUCGAUGUGAGCGUGU-UACCGACGGCAAAAGUCGUUGCAACGCCCACUACCUGGCUUAACCCGCCACUG---GCCGGGCGGGUCAUCU-UUUAUGAC ---(((.....))).(((...)))(((((((((((((-(..(((((((....))))))).))))).((.....)))))))(((((((....---....))))))))))).-........ ( -36.60, z-score = 0.22, R) >droSim1.chr3R 726171 111 + 27517382 ---GAGCAAAUCCCGCACGUUGUCGAGGUGGGCGUGU-UGCCGACGGCAAAAGUCGGUGCAACGCCCACUACCUGGCUUAACCCGCCACUG---GCCGGGCGGGUCAUCU-UUUAUGAC ---........(((((..........(((((((((..-.((((((.......))))))...)))))))))..((((((............)---))))))))))((((..-...)))). ( -46.70, z-score = -1.72, R) >droSec1.super_6 783641 111 + 4358794 ---GGGCAAAUCCCGCACGUGGUCGAGGUGGGCGUGU-UGCCGACGGCAAAAGUCGGUGCAACGCCCACUACCUGGCUUAACCCGCCACUG---GUCGGGCGGGUCAUCU-UUUAUGAC ---(((.....)))((.((.(((...(((((((((..-.((((((.......))))))...))))))))))))))))...(((((((....---....))))))).....-........ ( -50.00, z-score = -2.11, R) >droYak2.chr3R 1003282 105 + 28832112 ---GGGCAAAUCCCGCACGUUGUCGAGGUGGGCGUGU-UGCCGCCGGCAAAAGUCGGUGCAACGCCCACUACCUGGCUUAACCCG---------GUCGGUCGGGCCAUCU-UUUAUGAC ---(((.....))).......((((.(((((((((..-(((.((((((....))))))))))))))))))...(((((..(((..---------...)))..)))))...-....)))) ( -46.50, z-score = -2.08, R) >droEre2.scaffold_4770 985156 114 + 17746568 ---GGGCAAAUCCCGCACGUUGUCGAUGUGGGCGUGU-UGCCGUCGGCAAAAGUCGGUGAUACGCCCACUACCUGGCUUAACCCGCCACUGGCCAUCGGUCGGGCCCUCU-UUUAUGAC ---((((....................((((((((((-..(((.(.......).)))..))))))))))....((((.......))))((((((...))))))))))...-........ ( -48.30, z-score = -2.35, R) >droAna3.scaffold_13340 22897009 116 + 23697760 GACGGGUAAAUCCCGGACGUGGUCGAGGUGGGCGUGU-UGCCGACGGCAAAAGUCGGUGCAACGCCCACUACCUGGCUUAACCCGCCACCGGACGCUG--CGAGCCUCCUGUUUAUGAC ((((((....((.(((.((((((...(((((((((..-.((((((.......))))))...))))))))))))((((.......))))....))))))--.))....))))))...... ( -49.00, z-score = -1.57, R) >droPer1.super_3 2480380 112 - 7375914 ---AUGUAAAUCCCUUCGGC--UUGAGGUGGGCGUGCGUUCCGACGGCAAAAGUCGGUGCAACGCCCACC-CCUGGCUUAACCCGCCACCAUCCUACGCGCGGGCCUUCU-UUUAUGAC ---..(((((.......(((--(.(.(((((((((..((.(((((.......))))).)).)))))))))-.).))))...((((((..........).)))))......-)))))... ( -43.30, z-score = -2.09, R) >dp4.chr2 8679037 112 - 30794189 ---AAGUAAAUCCCUUCGGC--UUGAGGUGGGCGUGCGUUCCGACGGCAAAAGUCGGUGCAACGCCCACC-CCUGGCUUAACCCGCCACCAUCCUACGCGCGGGCCUUCU-UUUAUGAC ---..(((((.......(((--(.(.(((((((((..((.(((((.......))))).)).)))))))))-.).))))...((((((..........).)))))......-)))))... ( -43.40, z-score = -2.08, R) >consensus ___GGGCAAAUCCCGCACGUUGUCGAGGUGGGCGUGU_UGCCGACGGCAAAAGUCGGUGCAACGCCCACUACCUGGCUUAACCCGCCACUG___GCCGGGCGGGCCAUCU_UUUAUGAC ...........((((..((.......(((((((((...(((((((.......)))))))..)))))))))...((((.......))))........))..))))............... (-30.21 = -31.90 + 1.69)
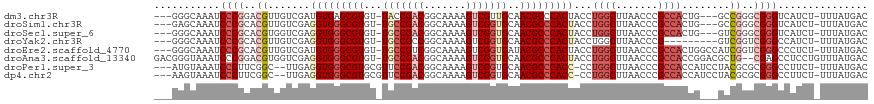
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:50:06 2011