
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,287,236 – 6,287,338 |
| Length | 102 |
| Max. P | 0.986212 |

| Location | 6,287,236 – 6,287,334 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 79.76 |
| Shannon entropy | 0.33123 |
| G+C content | 0.36082 |
| Mean single sequence MFE | -23.59 |
| Consensus MFE | -17.21 |
| Energy contribution | -17.64 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.986212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
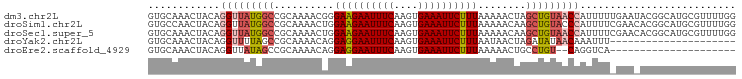
>dm3.chr2L 6287236 98 - 23011544 AAACUACAGGUUAUGGCCGCAAAACGGGAAGAAUUUCAAGUGAAAUUCUUUAAAAACUAGCUGUAACCAUUUUUGAAUACGGCAUGCGUUUUGGUUCA ........(((....))).(((((((.((((((((((....))))))))))........((((((..((....))..))))))...)))))))..... ( -28.30, z-score = -3.24, R) >droSim1.chr2L 6085329 98 - 22036055 CAACUACAGGUUAUGGCCGCAAAACUGGAAGAAUUUCAAGUGAAAUUCUUUAAAAACAAGCUGUACCCAUUUUCGAACACGGCAUGCGUUUUGGGUCA .............(((((.((((((..((((((((((....))))))))))........(((((..............)))))....))))))))))) ( -26.74, z-score = -2.22, R) >droSec1.super_5 4358488 98 - 5866729 AAACUACAGGUUAUGGCCGCAAAACUGGAAGAAUUUCAAGUGAAAUUCUUUAAAAACAAGCUGUAACCAUUUUCGAACACGGCAUGCGUUUUGGGUCA .............(((((.((((((..((((((((((....))))))))))........(((((..............)))))....))))))))))) ( -26.74, z-score = -2.50, R) >droYak2.chr2L 15703912 75 + 22324452 AAACUACAGGUUUUAGCCGCAAAACAGGAGGAAUUUCAAGUGAAAUUCUUUAAUAACUAGAUAUAACAAAUUUUA----------------------- ...(((..(((....))).........((((((((((....))))))))))......)))...............----------------------- ( -12.60, z-score = -2.09, R) >consensus AAACUACAGGUUAUGGCCGCAAAACUGGAAGAAUUUCAAGUGAAAUUCUUUAAAAACAAGCUGUAACCAUUUUCGAACACGGCAUGCGUUUUGGGUCA ........(((((((((..........((((((((((....))))))))))........))))))))).............................. (-17.21 = -17.64 + 0.44)


| Location | 6,287,240 – 6,287,338 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 77.06 |
| Shannon entropy | 0.38348 |
| G+C content | 0.38315 |
| Mean single sequence MFE | -21.56 |
| Consensus MFE | -14.79 |
| Energy contribution | -15.75 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.923909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
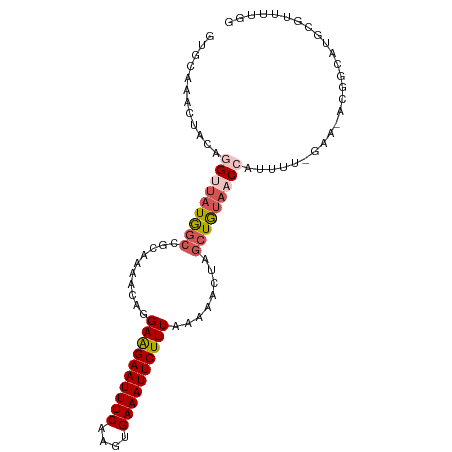
>dm3.chr2L 6287240 98 - 23011544 GUGCAAACUACAGGUUAUGGCCGCAAAACGGGAAGAAUUUCAAGUGAAAUUCUUUAAAAACUAGCUGUAACCAUUUUUGAAUACGGCAUGCGUUUUGG ............(((....))).(((((((.((((((((((....))))))))))........((((((..((....))..))))))...))))))). ( -28.30, z-score = -3.03, R) >droSim1.chr2L 6085333 98 - 22036055 GUGCCAACUACAGGUUAUGGCCGCAAAACUGGAAGAAUUUCAAGUGAAAUUCUUUAAAAACAAGCUGUACCCAUUUUCGAACACGGCAUGCGUUUUGG (.((((((.....))..))))).((((((..((((((((((....))))))))))........(((((..............)))))....)))))). ( -24.14, z-score = -1.23, R) >droSec1.super_5 4358492 98 - 5866729 GUGCAAACUACAGGUUAUGGCCGCAAAACUGGAAGAAUUUCAAGUGAAAUUCUUUAAAAACAAGCUGUAACCAUUUUCGAACACGGCAUGCGUUUUGG ((((........(((((((((..........((((((((((....))))))))))........))))))))).....((....))))))......... ( -25.07, z-score = -1.86, R) >droYak2.chr2L 15703914 77 + 22324452 GUGCAAACUACAGGUUUUAGCCGCAAAACAGGAGGAAUUUCAAGUGAAAUUCUUUAAUAACUAGAUAUAACAAAUUU--------------------- ((.....(((..(((....))).........((((((((((....))))))))))......))).....))......--------------------- ( -12.80, z-score = -1.58, R) >droEre2.scaffold_4929 15214755 75 - 26641161 GUGCAAACUACAGGUUAUAGCCGCAAAACAGGAGGAAUUUCAAGUGAAAUUCUUUAAAAACUGCCUGU--CAGGUCA--------------------- .......((((((((.....((........))(((((((((....)))))))))........))))))--.))....--------------------- ( -17.50, z-score = -1.42, R) >consensus GUGCAAACUACAGGUUAUGGCCGCAAAACAGGAAGAAUUUCAAGUGAAAUUCUUUAAAAACUAGCUGUAACCAUUUU_GAA_ACGGCAUGCGUUUUGG ............(((((((((..........((((((((((....))))))))))........))))))))).......................... (-14.79 = -15.75 + 0.96)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:20:58 2011