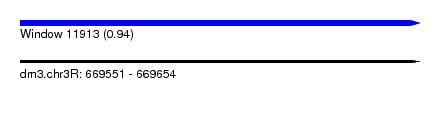
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 669,551 – 669,654 |
| Length | 103 |
| Max. P | 0.937173 |

| Location | 669,551 – 669,654 |
|---|---|
| Length | 103 |
| Sequences | 10 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 73.00 |
| Shannon entropy | 0.60133 |
| G+C content | 0.44097 |
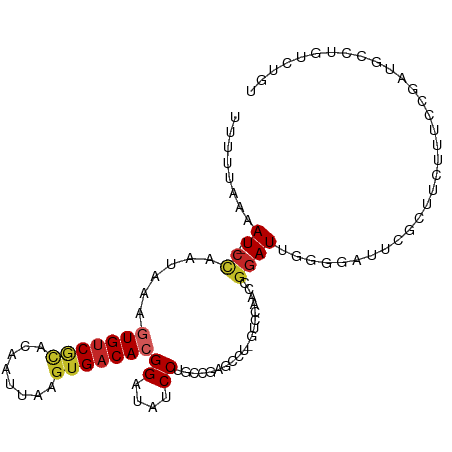
| Mean single sequence MFE | -27.67 |
| Consensus MFE | -9.58 |
| Energy contribution | -9.88 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.937173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
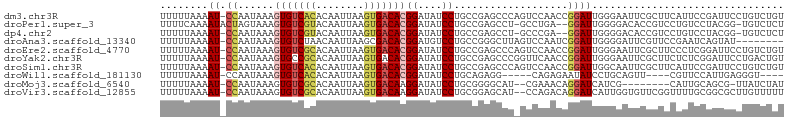
>dm3.chr3R 669551 103 + 27905053 UUUUUAAAAU-CCAAUAAAGUGUCACACAAUUAAGUGACACGGAUAUCCUGCCGAGCCCAGUCCAACCGGAUUGGGAAUUCGCUUCAUUCCGAUUCCUGUCUGU .......(((-(.......(((((((........)))))))(((......((.((.((((((((....))))))))...)))).....)))))))......... ( -31.50, z-score = -3.81, R) >droPer1.super_3 2470106 100 - 7375914 UUUUCAAAAUACUAGUAAAGUGUCGUACAAUUAAGUGACACGGAUAUCCUGCCGAGCCU-GCCUGA--GGAUUGGGGACACCGUCCUGUCCUACGG-UGUCUCU ...................((((((..........))))))((....))..((((.(((-.....)--)).))))(((((((((........))))-))))).. ( -27.60, z-score = -0.87, R) >dp4.chr2 8668715 99 - 30794189 UUUUUAAAAU-CCAAUAAAGUGUCGUACAAUUAAGUGACACGGAUAUCCUGCCGAGCCU-GCCCGA--GGAUUGGGGACACCGUCCUGUCCUACGG-UGUCUCU .......(((-((......((((((..........))))))((....))..........-......--)))))(((((((((((........))))-))))))) ( -29.50, z-score = -1.78, R) >droAna3.scaffold_13340 22887493 95 + 23697760 UUUUUAAAAU-CCAAUAAAGUGUCUAACAAUUAAGCGACACGGAUGUCCUGCCGGGCUUAGUCCAAUCGGAUUGGGGAUUCGUUCCGAAUCAGUAU-------- .......(((-((......(((((............)))))((((((((....))))...))))....)))))...((((((...)))))).....-------- ( -24.80, z-score = -1.43, R) >droEre2.scaffold_4770 974354 103 + 17746568 UUUUUAAAAU-CCAAUAAAGUGUCGCACAAUUAAGUGACACGGAUAUCCUGCCGAGCCCAGUCCAACCGGAUUGGGAAUUCGCUUCCCUCGGAUUCCUGUCUGU ..........-........(((((((........)))))))(((((.....(((((((((((((....))))))))...........))))).....))))).. ( -33.40, z-score = -3.65, R) >droYak2.chr3R 991623 103 + 28832112 UUUUUAAAAU-CCAAUAAAGUGCCGCACAAUUAAGUGACACGGAUAUCCUGCCGAGCCCGGUUCAACCGGAUUGGGAAUUCGCUUCUCUCGGAUUCCUGACUGU .......(((-((......(((.(((........))).)))((.......((((....))))....)))))))(((((((((.......)))))))))...... ( -26.50, z-score = -0.88, R) >droSim1.chr3R 712860 103 + 27517382 UUUUUAAAAU-CCAAUAAAGUGUCACACAAUUAAGUGACACGGAUAUCCUGCCGAGCCCAGUCCAACCGGAUUGGCAAUUCGCUUCAUUCCGAUUCCUGUCUGU .......(((-(.......(((((((........)))))))(((......((.((..(((((((....)))))))...)).)).....)))))))......... ( -27.80, z-score = -3.30, R) >droWil1.scaffold_181130 8530734 90 + 16660200 UUUUUAAAAU-CCAAUAAAGUGUCACACAAUUAAGUGACACGGAUAUCCUGCAGAGG-----CAGAGAAUAUCCUGCAGUU----CGUUCCAUUGAGGGU---- ........((-((......(((((((........)))))))(((((((((((....)-----))).).)))))).......----...........))))---- ( -26.50, z-score = -3.52, R) >droMoj3.scaffold_6540 23635249 92 + 34148556 UUUUUAAAAU-CCAAUAAAGUGUCGCACAAUUAAGUGACAAGGAUAUCCUGCGGGGCAU--CGAAACAGGAUCAUCG--------CAUUGCAGCG-UUAUCUAU ..........-.........((((((........)))))).(((((.(((((((.((((--(.......)))....)--------).)))))).)-.))))).. ( -19.80, z-score = -1.04, R) >droVir3.scaffold_12855 8422057 101 + 10161210 UUUUUAAAAU-CCAAUAAAGUGUCGCACAAUUAAGUGACAAGGAUAUCCUGCGGAGCAU--CCAGACAGGAUCAUUGGUGUUCGGUUUUGCGGCGCUUGUUUUU ..........-......((((((((((.((((....((((..((((((((((((.....--)).).)))))).)))..)))).)))).))))))))))...... ( -29.30, z-score = -2.21, R) >consensus UUUUUAAAAU_CCAAUAAAGUGUCGCACAAUUAAGUGACACGGAUAUCCUGCCGAGCCU_GUCCAACCGGAUUGGGGAUUCGCUUCUUUCCGAUGCCUGUCUGU ...........((......(((((((........)))))))((....))...................)).................................. ( -9.58 = -9.88 + 0.30)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:50:04 2011