
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 660,450 – 660,545 |
| Length | 95 |
| Max. P | 0.500000 |

| Location | 660,450 – 660,545 |
|---|---|
| Length | 95 |
| Sequences | 8 |
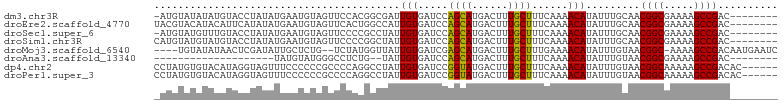
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 73.59 |
| Shannon entropy | 0.52979 |
| G+C content | 0.41769 |
| Mean single sequence MFE | -20.11 |
| Consensus MFE | -11.23 |
| Energy contribution | -10.85 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.18 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr3R 660450 95 + 27905053 -AUGUAUAUAUGUACCUAUAUGAAUGUAGUUCCACGGCGAUUGUGAUCCAGCAUGACUUUGCUUUCAAAACAUAUUUGCAACGGCGAAAAGCCGAC-------- -.((((.((((((..(((((....)))))..............(((...((((......)))).)))..)))))).)))).((((.....))))..-------- ( -22.00, z-score = -1.23, R) >droEre2.scaffold_4770 965776 96 + 17746568 UACGUACAUACAUUCAUAUAUGAAUGUAGUUCACUGGCCAUUGUGAUCCAGCAUGACUUUGCUUUCAAAACAUAUUUGCAACGGCGAAAAGCCGAC-------- ...(((..((((((((....))))))))..((((........))))...((((......)))).............)))..((((.....))))..-------- ( -22.00, z-score = -1.55, R) >droSec1.super_6 763982 95 + 4358794 -AUGUAUGUUUGUACCUAUAUGAAUGUAGUUCCCCGCCUAUUGUGAUCCAGCAUGACUUUGCUUUCAAAACAUAUUUGCAACGGCGAAAAGCCGAC-------- -..((((((((....(((((....)))))..............(((...((((......)))).)))))))))))......((((.....))))..-------- ( -19.20, z-score = -1.05, R) >droSim1.chr3R 703810 96 + 27517382 CAUGUAUGUAUGUACCUAUAUGAAUGUAGUUCCCCGGCUAUUGUGAUCCAGCAUGACUUUGCUUUCAAAACAUAUUUGCAACGGCGAAAAGCCGAC-------- ..((((.((((((............(((((......)))))..(((...((((......)))).)))..)))))).)))).((((.....))))..-------- ( -23.00, z-score = -1.59, R) >droMoj3.scaffold_6540 23626513 97 + 34148556 ----UGUAUAUAACUCGAUAUUGCUCUG--UCUAUGGUUAUUGUGAUCGAGCAUGACUUUGCUUUGAAAACAUAUUUGUAACGGC-AAAAGCCGACAAUGAAUC ----.....((((((.((((......))--))...))))))(((..(((((((......))).))))..)))...((((..((((-....))))))))...... ( -18.40, z-score = -0.37, R) >droAna3.scaffold_13340 22880361 74 + 23697760 --------------------UAUGUAUGGGCCUCUG--UAUUGUGAUCCAGCAUGACUUUGCUUUCAAAACAUAUUUGUAACGGCGAAAAGCCGAC-------- --------------------(((((.(((..(.(..--....).)..)))(((......))).......))))).......((((.....))))..-------- ( -17.10, z-score = -1.13, R) >dp4.chr2 8661480 98 - 30794189 CCUAUGUGUACAUAGGUAGUUUCCCCCCGCCCCAGGCCUAUUGUGAUCCGGUAUGACUUUGCUUUCAAAACAUAUUUGUAACGGCAAAAAGCCGACAC------ ((((((....))))))..((((......(((...)))......(((...((((......)))).)))))))..........((((.....))))....------ ( -19.60, z-score = 0.00, R) >droPer1.super_3 2462834 98 - 7375914 CCUAUGUGUACAUAGGUAGUUUCCCCCCGCCCCAGGCCUAUUGUGAUCCGGUAUGACUUUGCUUUCAAAACAUAUUUGUAACGGCAAAAAGCCGACAC------ ((((((....))))))..((((......(((...)))......(((...((((......)))).)))))))..........((((.....))))....------ ( -19.60, z-score = 0.00, R) >consensus _AUGUAUGUAUAUACCUAUAUGAAUGUAGUUCCACGGCUAUUGUGAUCCAGCAUGACUUUGCUUUCAAAACAUAUUUGCAACGGCGAAAAGCCGAC________ .........................................(((.....((((......))))......))).........((((.....)))).......... (-11.23 = -10.85 + -0.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:50:03 2011