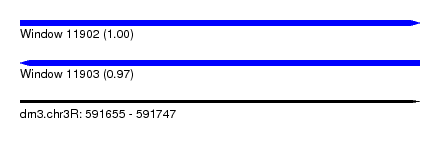
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 591,655 – 591,747 |
| Length | 92 |
| Max. P | 0.996423 |

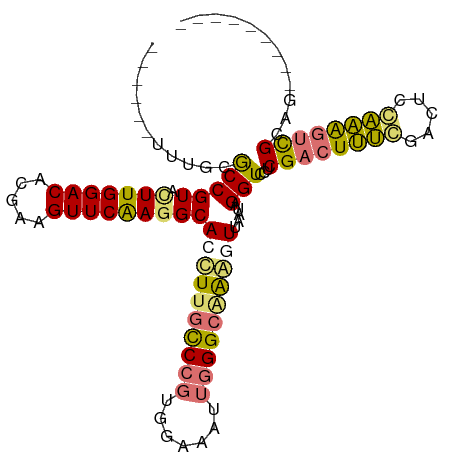
| Location | 591,655 – 591,747 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 69.62 |
| Shannon entropy | 0.45930 |
| G+C content | 0.53551 |
| Mean single sequence MFE | -35.75 |
| Consensus MFE | -27.04 |
| Energy contribution | -27.10 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.93 |
| SVM RNA-class probability | 0.996423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 591655 92 + 27905053 --------UUCGCCGUAUUUGGACACGAAGUUCGAGGCACCUUGCCCGUGGAAAUUGGGCAGAAUUAAUAGGUCCUCGACUUUCCACUCGAAAGUCGCAG--------- --------...((.......((((.....))))((((.((((((((((.......))))))........))))))))(((((((.....)))))))))..--------- ( -31.30, z-score = -2.34, R) >droSim1.chr3R 633440 94 + 27517382 ------UUUGCGCCGUACUUGGACACGAAGUUCAAGGCACCUUACCCGUGGAAAUUGGGCGAGGUUAAUAGGUCCUCGACUUUCGACUCCAAAGUCGCAG--------- ------..((((((((.(((((((.....)))))))))((((..((((.......))))..)))).....)))....((((((.......))))))))).--------- ( -30.90, z-score = -1.70, R) >droSec1.super_6 692323 94 + 4358794 ------UUCGCACCGUACUUGGACACGAAGUUCAAGGCACCUUGCCCGUGGAAAUUGGGCGAGGUUAAUAGGUCCUCGACUUUCGACUCGAAAGUCGCAG--------- ------...(((((((.(((((((.....)))))))))((((((((((.......)))))))))).....)))....((((((((...))))))))))..--------- ( -40.10, z-score = -4.18, R) >droAna3.scaffold_13340 22808202 109 + 23697760 CUCUCGAGUUCGCCGUACUUGGACGUCAAGUUCGAAGCACUUGGUCAAUGCAAAUGAGGCAAACUGGAGAGGUCCUCGACUCCGGGCCCCGGGCCUGGGUUCAUCCCAU (((((.((((.(((....((((((.....)))))).(((.((....)))))......))).)))).)))))......((..(((((((...)))))))..))....... ( -40.70, z-score = -1.19, R) >consensus ______UUUGCGCCGUACUUGGACACGAAGUUCAAGGCACCUUGCCCGUGGAAAUUGGGCAAAGUUAAUAGGUCCUCGACUUUCGACUCCAAAGUCGCAG_________ ...........(((((.(((((((.....)))))))))((((((((((.......)))))))))).....)))...((((((((.....))))))))............ (-27.04 = -27.10 + 0.06)

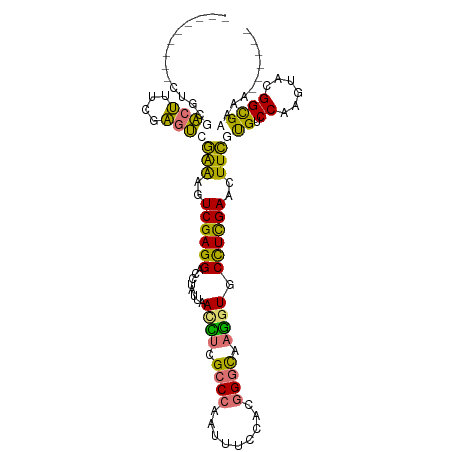
| Location | 591,655 – 591,747 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 69.62 |
| Shannon entropy | 0.45930 |
| G+C content | 0.53551 |
| Mean single sequence MFE | -34.98 |
| Consensus MFE | -18.76 |
| Energy contribution | -17.95 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.973477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 591655 92 - 27905053 ---------CUGCGACUUUCGAGUGGAAAGUCGAGGACCUAUUAAUUCUGCCCAAUUUCCACGGGCAAGGUGCCUCGAACUUCGUGUCCAAAUACGGCGAA-------- ---------.((((((((((.....)))))))((((((((........(((((.........))))))))).))))......(((((....))))))))..-------- ( -31.30, z-score = -2.27, R) >droSim1.chr3R 633440 94 - 27517382 ---------CUGCGACUUUGGAGUCGAAAGUCGAGGACCUAUUAACCUCGCCCAAUUUCCACGGGUAAGGUGCCUUGAACUUCGUGUCCAAGUACGGCGCAAA------ ---------.((((..((((((..((((..((((((........((((..(((.........)))..)))).))))))..))))..)))))).....))))..------ ( -34.60, z-score = -2.85, R) >droSec1.super_6 692323 94 - 4358794 ---------CUGCGACUUUCGAGUCGAAAGUCGAGGACCUAUUAACCUCGCCCAAUUUCCACGGGCAAGGUGCCUUGAACUUCGUGUCCAAGUACGGUGCGAA------ ---------...(((((((((...))))))))).(.(((.....((((.((((.........)))).))))..((((.((.....)).))))...))).)...------ ( -32.20, z-score = -2.03, R) >droAna3.scaffold_13340 22808202 109 - 23697760 AUGGGAUGAACCCAGGCCCGGGGCCCGGAGUCGAGGACCUCUCCAGUUUGCCUCAUUUGCAUUGACCAAGUGCUUCGAACUUGACGUCCAAGUACGGCGAACUCGAGAG .((((.....))))(((((((...)))).)))......(((((.((((((((......(((((.....)))))...(.(((((.....))))).))))))))).))))) ( -41.80, z-score = -1.94, R) >consensus _________CUGCGACUUUCGAGUCGAAAGUCGAGGACCUAUUAACCUCGCCCAAUUUCCACGGGCAAGGUGCCUCGAACUUCGUGUCCAAGUACGGCGAAAA______ .............((((....))))(((..((((((........((((.((((.........)))).)))).))))))..)))(((.((......)))))......... (-18.76 = -17.95 + -0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:49:55 2011