
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 578,502 – 578,602 |
| Length | 100 |
| Max. P | 0.500000 |

| Location | 578,502 – 578,602 |
|---|---|
| Length | 100 |
| Sequences | 13 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 72.99 |
| Shannon entropy | 0.54277 |
| G+C content | 0.48858 |
| Mean single sequence MFE | -28.53 |
| Consensus MFE | -14.91 |
| Energy contribution | -14.70 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.55 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr3R 578502 100 - 27905053 -CGGAGUUGCCAGCUUCUGGUCCAGCUGGUCCCACCUCUACAUGGCCAUCUCGUGGCCAACUGUG---AUACUAAUCUAUCUGUGGCAGUGAGUAAUCAAAAAC----------------- -.((.(..(((((((........)))))))..).))..(((.(((((((...)))))))(((((.---(((..........))).)))))..))).........----------------- ( -30.90, z-score = -1.67, R) >droGri2.scaffold_15074 4920034 93 - 7742996 -GGGUGUGGCUAGUUUCUGGUCCAGUUGGUCGCACCUCUAUAUGGCCAUCUCCUGGCCCACAGUG---AUACUCAUCUAUUUGUGGCAGUAAGUAAC------------------------ -((((((((((((...(((...)))))))))))))))......(((((.....)))))(((((.(---((....)))...)))))............------------------------ ( -28.10, z-score = -0.63, R) >droMoj3.scaffold_6540 23542865 100 - 34148556 -AGGAUUGGCCAGCUUUUGGUCUAGCUGGUCGCAUCUUUAUAUGGCCAUUUCUUGGCCCACAGUA---AUACUCAUUUACCUUUGGCAGUAAGUAUUAAUAAAC----------------- -((((((((((((((........))))))))).))))).....(((((.....))))).....((---(((((.(((..((...)).))).)))))))......----------------- ( -29.30, z-score = -2.28, R) >droVir3.scaffold_12855 8318552 117 - 10161210 -AGGCGUUGCCAGUUUUUGGUCCAGCUGGUCUCACCUCUAUAUGGCCAUCUCCUGGCCCACAGUC---AUACUCAUCUACCUGUGGCAGUAAGUAUCAAAGUCUUAUUGGUUAGUGAGAAC -.((((..(((((((........)))))))..)....(.....)))).(((((((((((((((..---............))))))(((((((.........)))))))))))).)))).. ( -33.24, z-score = -1.08, R) >droAna3.scaffold_13340 22799021 80 - 23697760 UGGGC-UGGCUAGCUUCUUUUCCAGCUGGUCCCACCUCUACAUGGCCAUCUCCUGGCCAACUGUG---AUACUUAUCUAUCUGU------------------------------------- .((..-.((((((((........))))))))...))..((((((((((.....))))))..))))---................------------------------------------- ( -23.60, z-score = -1.77, R) >droSim1.chr3R 626043 88 - 27517382 -CGGAGUGGCCAGCUUCUGGUCCAGCUGGUCCCACCUCUACAUGGCCAUCUCGUGGCCAACUGUG---AUACUUAUCUAUCUGUGGCAGUGA----------------------------- -.(((((((((((((........))))))..))).))))...(((((((...)))))))(((((.---(((..........))).)))))..----------------------------- ( -31.50, z-score = -1.25, R) >droEre2.scaffold_4770 890295 97 - 17746568 -CGGAGUGGCCAGCUUCUGGUCCAGCUGGUCCCACCUCUACAUGGCCAUCUCGUGGCCAACUGUG---AUACUAAUCUAUCUGUGGCAGUAAGUAAUCGAA-------------------- -.((.(.((((((((........)))))))).).))..(((.(((((((...)))))))(((((.---(((..........))).)))))..)))......-------------------- ( -32.50, z-score = -1.67, R) >droYak2.chr3R 895721 97 - 28832112 -CGGAGUAGCCAGCUUUUGGUCCAGCUGGUCCCACCUCUACAUGGCCAUCUCGUGGCCAACUGUG---AUACUAAUUUAUCUGUGGCAGUAAGUAAUCAAA-------------------- -.((.(..(((((((........)))))))..).))..(((.(((((((...)))))))(((((.---(((..........))).)))))..)))......-------------------- ( -31.10, z-score = -2.15, R) >droWil1.scaffold_181130 1545642 98 + 16660200 -GGGCGUUGCAAGUUUCUGGUCCAGUUGGUCACAUCUUUAUAUGGCCAUUUCAUGGCCCACUGUA---AUACUUAUCUAUCUAUGGCAGUAAGCAUGGAAAA------------------- -(((..(((((.((....((.(((..((((((.((....)).)))))).....))))).))))))---)..))).....((((((........))))))...------------------- ( -21.70, z-score = 0.50, R) >droSec1.super_6 685424 100 - 4358794 -CGGAGUGGCCAGCUUCUGGUCCAGCUGGUCCCACCUCUACAUGGCCAUCUCGUGGCCAACUGUG---AUACUUAUCUAUCUGUGGCAGUGAGUAAUCAAAAAC----------------- -.(((((((((((((........))))))..))).))))...(((((((...)))))))(((((.---(((..........))).)))))((....))......----------------- ( -32.60, z-score = -1.66, R) >dp4.chr2 8579324 101 + 30794189 -GGGCGUGGCAAGCUUUUGGUCCAGCUGGUCGCAUCUCUACAUGGCCAUCUCCUGGCCAACAGUG---AUACUGAUCUACCUGUGGCAGUGAGUACUACAAAGAC---------------- -....((((...((((...((((((..(((((.(((.((...((((((.....))))))..)).)---))..)))))...))).)))...)))).))))......---------------- ( -29.10, z-score = 0.60, R) >droPer1.super_3 2379921 101 + 7375914 -GGGCGUGGCAAGCUUUUGGUCCAGCUGGUCGCAUCUCUACAUGGCCAUCUCCUGGCCAACAGUG---AUACUGAUCUACCUGUGGCAGUGAGUACUACAAAGAC---------------- -....((((...((((...((((((..(((((.(((.((...((((((.....))))))..)).)---))..)))))...))).)))...)))).))))......---------------- ( -29.10, z-score = 0.60, R) >apiMel3.Group12 5180739 100 - 9182753 -GGGUAUAGCUUAUUUGUUACGCGAAUGGAGACACAUUCAAUUAGCGAUAUCUUUGCCUGCUCUGCUUCUAUUAAUACAUCUAUG---GUAAGUAUACAUAAUU----------------- -(((((.((.....((((((...(((((......)))))...))))))...)).)))))....(((((((((..........)))---).))))).........----------------- ( -18.10, z-score = -0.04, R) >consensus _GGGAGUGGCCAGCUUCUGGUCCAGCUGGUCCCACCUCUACAUGGCCAUCUCCUGGCCAACUGUG___AUACUAAUCUAUCUGUGGCAGUAAGUAAUCAAAA___________________ ..((....(((((((........)))))))....)).(((((.(((((.....)))))....(((....))).........)))))................................... (-14.91 = -14.70 + -0.21)
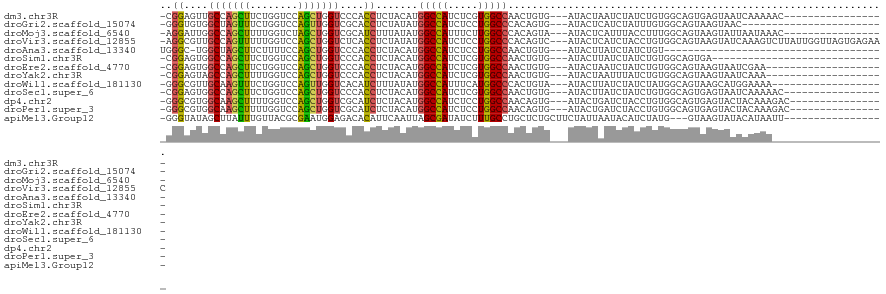

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:49:54 2011