| Sequence ID | dm3.chr3R |
|---|---|
| Location | 561,326 – 561,429 |
| Length | 103 |
| Max. P | 0.995195 |

| Location | 561,326 – 561,429 |
|---|---|
| Length | 103 |
| Sequences | 13 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 62.69 |
| Shannon entropy | 0.79395 |
| G+C content | 0.42492 |
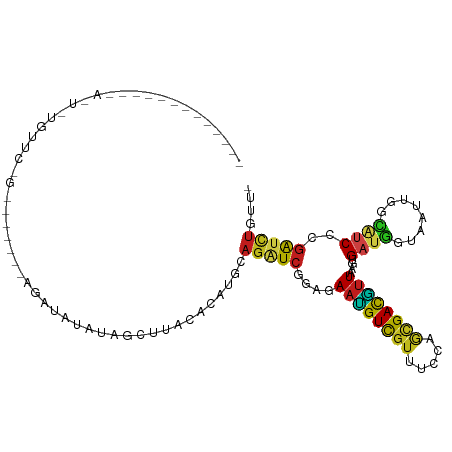
| Mean single sequence MFE | -23.85 |
| Consensus MFE | -12.72 |
| Energy contribution | -12.09 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.76 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.995195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
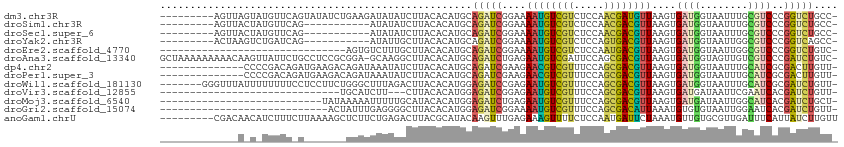
>dm3.chr3R 561326 103 + 27905053 ---------AGUUAGUAUGUUCAGUAUAUCUGAAGAUAUAUCUUACACAUGCAGAUCGGAAAAUGUCGUCUCCAACGAUGUUAAGUGAUGGUAAUUUGCGUCCCGGUCUGCC- ---------.....((((.(((((.....))))).))))...........(((((((((..((((((((.....))))))))....((((........))))))))))))).- ( -31.60, z-score = -3.49, R) >droSim1.chr3R 609387 92 + 27517382 ---------AGUUACUAUGUUCAG-----------AUAUAUCUUACACAUGCAGAUCGGAAAAUGUCGUCUCCAACGACGUUAAGUGAUGGUAAUUUGCGUCCCGGUCUGCC- ---------.......((((..((-----------(....)))...))))(((((((((..((((((((.....))))))))....((((........))))))))))))).- ( -27.30, z-score = -2.64, R) >droSec1.super_6 668476 92 + 4358794 ---------AGUUACUAUGUUCAG-----------AUAUAUCUUACACAUGCAGAUCGGAAAAUGUCGUCUCCAACGACGUUAAGUGAUGGUAAUUUGCGUCCCGGUCUGCC- ---------.......((((..((-----------(....)))...))))(((((((((..((((((((.....))))))))....((((........))))))))))))).- ( -27.30, z-score = -2.64, R) >droYak2.chr3R 876785 92 + 28832112 ---------ACUAAGUCUGAUCAG-----------AUAUUGCUUACACAUGCAGAUCGGAAAAUGUCGUCUCCAGUGACGUUAAGUGAUGGUAAUUGGCGUCCCGGUCAGCC- ---------.....(.((((((.(-----------((.((((........)))))))((...((((((...(((.(.((.....)).))))....)))))).))))))))).- ( -22.90, z-score = -0.34, R) >droEre2.scaffold_4770 872005 81 + 17746568 -------------------------------AGUGUCUUUGCUUACACAUGCAGAUCGGAAAAUGUCGUCUCCAAUGACGUUAAGUGAUGGUAAUUGGCGUCCCGGUCUGUC- -------------------------------.((((........))))..(((((((((..((((((((.....))))))))....((((.(....).))))))))))))).- ( -25.20, z-score = -2.47, R) >droAna3.scaffold_13340 22785309 111 + 23697760 GCUAAAAAAAAACAAGUUAUUCUGCCUCCGCGGA-GCAAGGCUUACACAUGCAGAUCUGAGAAUGUCGAUUCCAGCGACGUUAAGUGAUGGUAGUUGUCGUCCCGAUCUGUC- .............(((((.((((((....)))))-)...)))))......(((((((....(((((((.......)))))))..(.(((((......))))).)))))))).- ( -26.70, z-score = -0.28, R) >dp4.chr2 24138920 98 + 30794189 --------------CCCCGACAGAUGAAGACAGAUAAAUAUCUUACACAUGCAGAUCGAAGAACGUCGUUUCCAGCGACGUUAAGUGAUGGUAAUUUGCAUCGCGACUUGUU- --------------...((...((((.....((((....))))......(((..((((...((((((((.....))))))))...)))).))).....)))).)).......- ( -22.20, z-score = -0.85, R) >droPer1.super_3 6910936 98 + 7375914 --------------CCCCGACAGAUGAAGACAGAUAAAUAUCUUACACAUGCAGAUCGAAGAACGUCGUUUCCAGCGACGUUAAGUGAUGGUAAUUUGCAUCGCGACUUGUU- --------------...((...((((.....((((....))))......(((..((((...((((((((.....))))))))...)))).))).....)))).)).......- ( -22.20, z-score = -0.85, R) >droWil1.scaffold_181130 1531167 105 - 16660200 -------GGGUUUAUUUUUUUUCCUCCUUCUGGGCUUUAGACUUACACAUGGAGAUCCGAGAAUGUCGUUUCCAGCGACGUUAAGUGAUGGUAAUUUGCAUCGCGAUCUGUU- -------(((((((.......(((.......)))...)))))))........(((((....((((((((.....))))))))..((((((........)))))))))))...- ( -25.00, z-score = -0.25, R) >droVir3.scaffold_12855 8302182 79 + 10161210 ------------------------------UGCAUCUU---CUUACACAUGGAGAUCGGAGAAUGUCGUUUCCAGCGACGUUAAGUGAUGAUAAUUCGAAUCACGAUCUGUU- ------------------------------...(((((---(........))))))((((.((((((((.....))))))))..(((((..........)))))..))))..- ( -20.70, z-score = -0.83, R) >droMoj3.scaffold_6540 23526391 85 + 34148556 ---------------------------UAUAAAAAUUUUUGCAUACACAUGGAGAUCUGAGAAUGUCGUUUCCAGCGACGUUAAGUGAUGAUAAUUGGCAUCACGAUCUGCU- ---------------------------.......................(.(((((....((((((((.....))))))))..((((((........))))))))))).).- ( -23.40, z-score = -2.35, R) >droGri2.scaffold_15074 4902165 84 + 7742996 ----------------------------ACUAUUUGAGGGGCUUACACAUGGAGAUCGGAAAAUGUCGUUUCCAGCGACAUUAAAUGUGUGUAAUUGGAAUCACGAUCUGUU- ----------------------------.((((.((.........)).))))((((((((.....))(.((((((..((((.......))))..)))))).).))))))...- ( -17.30, z-score = 0.16, R) >anoGam1.chrU 51762835 104 - 59568033 ---------CGACAACAUCUUUCUUAAAAGCUCUUCUGAGACUUACGCAUACAAGUUUGAGAAAGUUUUCUCCAAUGAUUCUAAAUGUUGUGCGUUGAUUUCAUUAUCUUGUU ---------.(((((...........((((((.((((.((((((........)))))).))))))))))....(((((.....((((.....))))....)))))...))))) ( -18.20, z-score = -0.28, R) >consensus _____________A_U_UGUUC_G_______AGAUAUAUAGCUUACACAUGCAGAUCGGAGAAUGUCGUUUCCAGCGACGUUAAGUGAUGGUAAUUGGCAUCCCGAUCUGUU_ ....................................................(((((....((((((((.....))))))))....((((........))))..))))).... (-12.72 = -12.09 + -0.63)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:49:51 2011