
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 534,597 – 534,717 |
| Length | 120 |
| Max. P | 0.985833 |

| Location | 534,597 – 534,717 |
|---|---|
| Length | 120 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.65 |
| Shannon entropy | 0.34343 |
| G+C content | 0.60139 |
| Mean single sequence MFE | -49.88 |
| Consensus MFE | -38.12 |
| Energy contribution | -37.36 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.985833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 534597 120 - 27905053 CUUAUCCUCGAUGGACUGCAUCUUGGCAUUGGCCGUUGCUGUGGCUUCGCUCAAUGAUUGCGUGGCCGCCGCUGCGGCCGUUGCUGAUUCGGUCCAGGACUCGGCGGAGUUUCCUUUUAC ....((((((((((((((.(((..((((.(((((((.((.((((((.(((.........))).)))))).)).))))))).))))))).)))))))....)))).)))............ ( -56.00, z-score = -4.41, R) >droSim1.chr3R 587040 120 - 27517382 CUUAUCCUCGAUGGACUGCAUUUUGGCAUUGGCCGUUGCUGUGGCUUCGCUCAAUGAUUGUGUGGCCGCCGCUGCGGCCGUCGCUGAUUCGGUCCAGGACUCGGCGGAGUUUCCCUUGAC ....((((((((((((((....(..((..(((((((.((.((((((.(((.........))).)))))).)).)))))))..))..)..)))))))....)))).)))............ ( -51.70, z-score = -2.86, R) >droSec1.super_6 647143 120 - 4358794 CUUAUCCUCGAUGGACUGCAUCUUGGCAUUGGCCGUUGCGGUGGCUUCGCUCAAUGAUUGUGUGGCCGCCGCUGCGGCCGUGGCUGAUUCGGUCCAGGACUCGGCGGAGUUUCCCUUGAC ....((((((((((((((.(((..(.(((.((((((.(((((((((.(((.........))).))))))))).))))))))).).))).)))))))....)))).)))............ ( -58.20, z-score = -3.95, R) >droYak2.chr3R 853256 120 - 28832112 CUUAUCCUCAAUGGACUGCAUCUUGGCAUUGGCCGUUGCUGUAGCUUCGCUCAAAGAUUGGGUGGCCGCAGCUGCGGCCGUCGCUGAUUCUGUCCAGGACUCGGCCGAGUUUCCCCUGAC .......(((..(((......((((((...((((((.((((..((..((((((.....))))))))..)))).))))))(((.(((........))))))...))))))..)))..))). ( -48.30, z-score = -2.19, R) >droEre2.scaffold_4770 846021 120 - 17746568 CUUAUCCUCGAUGGACUGCAUCUUGGCGUUGGCCGUUGCUGUAGCUUCGCUCAAAGAUUGGGUGGCCGCAGCUGCGGCCGUCGCUGAUUCUGUCCAGGACUCGGCCGAGUUUCCCUUGGC ....((((.((((((....(((..((((.(((((((.((((..((..((((((.....))))))))..)))).))))))).))))))))))))).))))....((((((.....)))))) ( -53.90, z-score = -3.03, R) >droAna3.scaffold_13340 22760392 120 - 23697760 UUUCUCCUCGAUGGACAGCAUCUUGGCGUUCGCCGUGGCGGUGGCUUCGCUCAGGGACUGAGUGGCCGCCGCAGCCGCCGUCGCCGAUUCCGUCCAGGACUCGGCCGAGUUGCCCUUAAC ......((((((((.(.((.....(((....)))...((((((((..(((((((...))))))))))))))).)).))))))(((((.(((.....))).))))).)))........... ( -56.70, z-score = -2.25, R) >droPer1.super_3 6887677 120 - 7375914 CUUCUCCUCGAUGGACUGCAUCUUGGCAUUGGCCGUCGCCGUGGCUUCGCUCAGCGAUUGUGUGGCCGCGGCCGCCGCCGUUGCCGAUUCUGUCCAGGACUCGGCCGAGUUUCCUUUCAC .........(((((((.(.(((..((((.((((.((.(((((((((.(((.........))).))))))))).)).)))).))))))).).)))))((((((....))))))....)).. ( -51.10, z-score = -2.32, R) >dp4.chr2 24110978 120 - 30794189 CUUCUCCUCGAUGGACUGCAUCUUGGCAUUGGCCGUCGCCGUGGCUUCGCUCAGCGAUUGUGUGGCCGCGGCCGCCGCCGUUGCCGAUUCUGUCCAGGACUCGGCCGAGUUUCCUUUCAC .........(((((((.(.(((..((((.((((.((.(((((((((.(((.........))).))))))))).)).)))).))))))).).)))))((((((....))))))....)).. ( -51.10, z-score = -2.32, R) >droWil1.scaffold_181130 1507423 120 + 16660200 CUUCUCCUCAAUGGACUGCAUCUUGGCAUUUGCUGUGGCUGUGGCUUCGCUUAGCGAUUGGGUGGCCGCUGCCGCUGCAGUGGCGGAUUCUGUCCACGAUUCGGCUGAAUUUCCUUUGGC ...........(((((.(.((((.(.((((.((.(((((.((((((((((...))))......)))))).))))).)))))).))))).).))))).......((..((.....))..)) ( -42.00, z-score = -0.57, R) >droVir3.scaffold_13047 6480505 120 - 19223366 CUUCUCCUCAAUGGACAGCAUUUUUGCAUUGGCCGUGGCGGUUGCCUCGCUUAGCGAUUGGGUGGCGGCCGCUGCCGCCGUUGCAGAUUCCGUCCAUGAUUCCGCCGAGUUGCCCUUCAC ..........((((((.(....((((((.((((.(..(((((((((((((...))))......)))))))))..).)))).))))))..).))))))(((((....)))))......... ( -47.30, z-score = -1.74, R) >droMoj3.scaffold_6540 32552979 120 + 34148556 CUUCUCUUCAAUAGACAACAUCUUUGCAUUGGCAGUUGCGGUUGCCUCACUCAGCGAUUGUGUUGCGGCCGCUGCCGCCGUAGCCGAUUCCGUCCACGACUCAGCUGAGGUGCCUUUGAC .......((((.((....((((((.((...(((.((.((((((((..(((.........)))..)))))))).)).)))((.(.((....)).).))......)).)))))).)))))). ( -36.50, z-score = -0.25, R) >droGri2.scaffold_14906 11783481 120 - 14172833 CUUCUCCUCAAUGGACAGCAUCUUCGCAUUGGCCGUGGCAGUUGCCUCGCUCAGCGAUUGAGUUGCGGCCGCUGCCGCUGUUGCCGAUUCCGUCCACGACUCCGCUGAGUUGCCCUUUAC ...........(((((.((......))((((((.((((((((.(((.(((((((...))))).)).))).))))))))....))))))...)))))((((((....))))))........ ( -45.70, z-score = -2.48, R) >consensus CUUCUCCUCGAUGGACUGCAUCUUGGCAUUGGCCGUUGCUGUGGCUUCGCUCAGCGAUUGUGUGGCCGCCGCUGCCGCCGUUGCCGAUUCUGUCCAGGACUCGGCCGAGUUUCCCUUGAC ...........(((((.(.(((..(((..((((.((.((((((((..(((.(((...))).))))))))))).)).))))..)))))).).)))))((((((....))))))........ (-38.12 = -37.36 + -0.76)
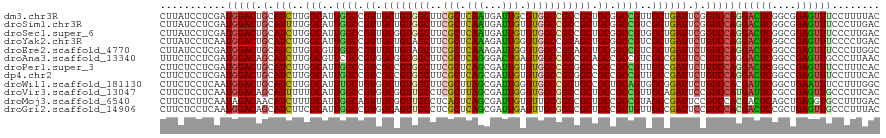

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:49:47 2011