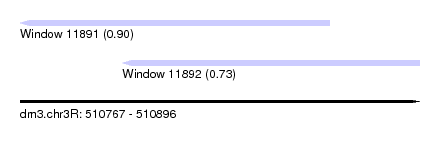
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 510,767 – 510,896 |
| Length | 129 |
| Max. P | 0.899618 |

| Location | 510,767 – 510,867 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 72.85 |
| Shannon entropy | 0.49270 |
| G+C content | 0.51021 |
| Mean single sequence MFE | -23.49 |
| Consensus MFE | -9.68 |
| Energy contribution | -10.56 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.899618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
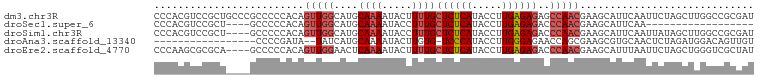
>dm3.chr3R 510767 100 - 27905053 CCCACGUCCGCUGCCCGCCCCCACAGUUGGCAUGCAAAAUACUUUUGCUCUCAUACCUUGAGAGAGCCAACGAAGCAUUCAAUUCUAGCUUGGCCGCGAU ........(((.(((..........((((((..(((((.....)))))(((((.....)))))..)))))).((((...........))))))).))).. ( -30.80, z-score = -3.28, R) >droSec1.super_6 623363 78 - 4358794 CCCACGUCCGCU----GCCCCCACAGUUGGCAUGCAAAAUACCUUUGCUCUCAUACCUUGAGAGACCCAACGAAGCAUUCAA------------------ .....(((.(((----(......)))).)))((((............((((((.....))))))..........))))....------------------ ( -17.15, z-score = -2.34, R) >droSim1.chr3R 562262 96 - 27517382 CCCACGUCCGCU----GCCCCCACAGUUGGCAUGCAAAAUACCUUUGCUCUCAUACCUUGAGAGACCCAACGAAGCAUUCAAUUAUAGCUUGGCCGCGAU ........(((.----(((......(((((...(((((.....)))))(((((.....)))))...))))).((((...........))))))).))).. ( -26.40, z-score = -3.05, R) >droAna3.scaffold_13340 22720291 80 - 23697760 -----------------CCCCGAUA--UAUCAUGCAAAAUACUUGUG-UCCCAUACCUUGGGAGAACCAGCGAAGCGUGCAACUCUAGAUGGACAGUUGU -----------------........--...(((((......((.((.-(((((.....)))))..)).))....)))))(((((((....)))..)))). ( -14.90, z-score = 0.46, R) >droEre2.scaffold_4770 820886 96 - 17746568 CCCAAGCGCGCA----GCCCCCACAGUUGGAACUCAAAAUACUUUUGCUCUCAUACCUUGAGAGACCCAACGAAGCAUUUAAUUCUAGCUGGGUCGCUAU ....(((((.((----((.......(((((....((((.....))))((((((.....))))))..)))))(((........)))..)))).).)))).. ( -28.20, z-score = -3.23, R) >consensus CCCACGUCCGCU____GCCCCCACAGUUGGCAUGCAAAAUACUUUUGCUCUCAUACCUUGAGAGACCCAACGAAGCAUUCAAUUCUAGCUGGGCCGCGAU .........................(((((....((((.....))))((((((.....))))))..)))))............................. ( -9.68 = -10.56 + 0.88)


| Location | 510,800 – 510,896 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 67.74 |
| Shannon entropy | 0.56642 |
| G+C content | 0.46412 |
| Mean single sequence MFE | -17.67 |
| Consensus MFE | -7.61 |
| Energy contribution | -8.00 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.728888 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 510800 96 - 27905053 AACUGCAACCUAAAGGUAUCUUCUCACUCCCCACGUCCGCUGCCCGCCCCCACAGUUGGCAUGCAAAAUACUUUUGCUCUCAUACCUUGAGAGAGC ....((....(((((((((......((.......))..((((((.((.......)).)))).))...)))))))))((((((.....)))))).)) ( -20.70, z-score = -1.58, R) >droSec1.super_6 623378 92 - 4358794 AACUGCAACCUAAAGGUAUCUGCUCACUCCCCACGUCCGCU----GCCCCCACAGUUGGCAUGCAAAAUACCUUUGCUCUCAUACCUUGAGAGACC ..........(((((((((.(((...........(((.(((----(......)))).)))..)))..)))))))))((((((.....))))))... ( -22.72, z-score = -2.88, R) >droSim1.chr3R 562295 92 - 27517382 AACUGCAACCUAAAGGUAUCUGCUCACUCCCCACGUCCGCU----GCCCCCACAGUUGGCAUGCAAAAUACCUUUGCUCUCAUACCUUGAGAGACC ..........(((((((((.(((...........(((.(((----(......)))).)))..)))..)))))))))((((((.....))))))... ( -22.72, z-score = -2.88, R) >droVir3.scaffold_13047 11794053 74 - 19223366 GGCUUCGGCUUAAGAGCUUAUUCAAAAUUAUUAAAAGCCCU----AUUUUU-CAUCAAAAAUGCAUAAAAUUAUGAAUG----------------- (((....)))........((((((.((((.(((...((...----((((((-....)))))))).))))))).))))))----------------- ( -7.50, z-score = 0.79, R) >droEre2.scaffold_4770 820919 91 - 17746568 AACUGCAACCUAAA-GUAUCUUCUUACUCCCCAAGCGCGCA----GCCCCCACAGUUGGAACUCAAAAUACUUUUGCUCUCAUACCUUGAGAGACC ...........(((-((((.((((.(((......((.....----))......))).))))......)))))))..((((((.....))))))... ( -14.70, z-score = -1.07, R) >consensus AACUGCAACCUAAAGGUAUCUUCUCACUCCCCACGUCCGCU____GCCCCCACAGUUGGCAUGCAAAAUACUUUUGCUCUCAUACCUUGAGAGACC ..........(((((((((...................((.....))..((......))........)))))))))((((((.....))))))... ( -7.61 = -8.00 + 0.39)

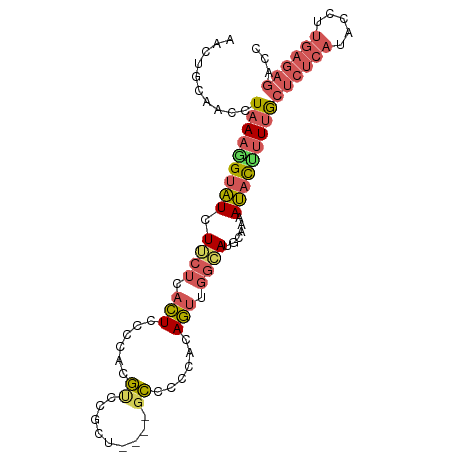

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:49:46 2011