| Sequence ID | dm3.chr3R |
|---|---|
| Location | 490,208 – 490,310 |
| Length | 102 |
| Max. P | 0.763405 |

| Location | 490,208 – 490,310 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 54.29 |
| Shannon entropy | 0.83602 |
| G+C content | 0.51738 |
| Mean single sequence MFE | -23.93 |
| Consensus MFE | -9.65 |
| Energy contribution | -9.47 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.73 |
| Mean z-score | -0.75 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.763405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
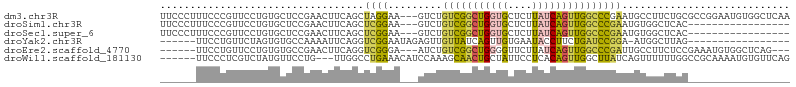
>dm3.chr3R 490208 102 + 27905053 UUCCCUUUCCCGUUCCUGUGCUCCGAACUUCAGCUAGGAA---GUCUGUCGGCUGGUGCUCUUAUCAGUUGGCCCGAAUGCCUUCUGCGCCGGAAUGUGGCUCAA ........((((((((.((((...((.((((......)))---))).(((((((((((....))))))))))).............)))).)))))).))..... ( -31.80, z-score = -1.29, R) >droSim1.chr3R 542358 85 + 27517382 UUCCCUUUCCCGUUCCUGUGCUCCGAACUUCAGCUCGGAA---GUCUGUCGGCUGGUGCUCUUAUCAGUUGGCCCGAAUGUGGCUCAC----------------- ........(((((((....(((((((.(....).)))).)---))..(((((((((((....)))))))))))..))))).)).....----------------- ( -23.90, z-score = -0.85, R) >droSec1.super_6 604037 85 + 4358794 UUCCCUUUCCCGUUCCUGUGCUCCGAACUUCAGCUCGGAA---GUCUGUCGGCUGGUGCUCUUAUCAGUUGGCCCGAAUGUGGCUCAC----------------- ........(((((((....(((((((.(....).)))).)---))..(((((((((((....)))))))))))..))))).)).....----------------- ( -23.90, z-score = -0.85, R) >droYak2.chr3R 806690 81 + 28832112 ------UUCCUGUUCUAGUGUGCCAAAAUUCAGGUCGGAAUAGAGUUGUUAUCAGUUGUGAAUACCUUCUGAUCCGGA-AUGGCUUAG----------------- ------...............((((...(((.((((((((.....(..(........)..).....)))))))).)))-.))))....----------------- ( -15.30, z-score = 0.22, R) >droEre2.scaffold_4770 799252 93 + 17746568 ------UUCCUGUUCCUGUGUGCCGAACUUCAGGUCGGGA---AUCUGUCGGCUGGGGUUCUUAUCAGUUGGCCCGAUUGCCUUCUCCGAAAUGUGGCUCAG--- ------.........(((...((((...(((.((..((.(---(((.(((((((((........)))))))))..)))).))....)))))...)))).)))--- ( -27.80, z-score = -0.32, R) >droWil1.scaffold_181130 1452466 96 - 16660200 ------UUCCCUCGUCUAUGUUCCUG---UUGGCCUGAAACAUCCAAAGCAACUGCUAUUCCUCACAGUUGGCUUAUCAGUUUUUUGGCCGCAAAAUGUGUUCAG ------.......(..((((((..((---(.((((.(((((.....(((((((((..........))))).))))....)))))..))))))).))))))..).. ( -20.90, z-score = -1.38, R) >consensus ______UUCCCGUUCCUGUGCUCCGAACUUCAGCUCGGAA___GUCUGUCGGCUGGUGCUCUUAUCAGUUGGCCCGAAUGUGGCUUAC_________________ ..................................((((.........(((((((((((....)))))))))))))))............................ ( -9.65 = -9.47 + -0.18)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:49:44 2011