
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 395,635 – 395,714 |
| Length | 79 |
| Max. P | 0.908581 |

| Location | 395,635 – 395,714 |
|---|---|
| Length | 79 |
| Sequences | 3 |
| Columns | 81 |
| Reading direction | forward |
| Mean pairwise identity | 66.80 |
| Shannon entropy | 0.45348 |
| G+C content | 0.42890 |
| Mean single sequence MFE | -20.27 |
| Consensus MFE | -13.77 |
| Energy contribution | -14.33 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.908581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 395635 79 + 27905053 UUUCAUUGCAAUCAUCCUGGGCUAAAUCUUCCGAUAAGCUUUGUCUUGCCAUUUCGGUGGAUGUGGCCUCAGGACCUUU-- ..............((((((((((.((((.((((...((........))....)))).)))).))))).))))).....-- ( -27.50, z-score = -3.38, R) >droSim1.chr3R_random 129097 79 + 1307089 UUUCAUUGCAAUCAUCCAGGGCUAAAUCUUCCGAUUAGCUUUGUCUUGCCAUUUCGGUGGAUGCGGCCGCAGGACCUUU-- ..............(((.(((((..((((.((((...((........))....)))).))))..)))).).))).....-- ( -21.20, z-score = -0.61, R) >droWil1.scaffold_180777 2693156 80 + 4753960 UCUCUUUUUAGGCUUCUUGGGGCAAAGUUUCAAAUAUUUUAUUCCUUGCCAUUUUUGUUAACUUGGC-GUAAAUCUUCGCU ..........(((.....((((.(((((.......))))).))))..)))..............(((-(........)))) ( -12.10, z-score = -0.37, R) >consensus UUUCAUUGCAAUCAUCCUGGGCUAAAUCUUCCGAUAAGCUUUGUCUUGCCAUUUCGGUGGAUGUGGCCGCAGGACCUUU__ ..............((((((((((.(((..((((...((........))....))))..))).))))).)))))....... (-13.77 = -14.33 + 0.57)

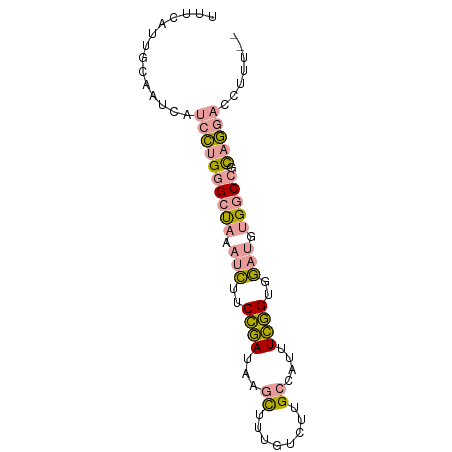
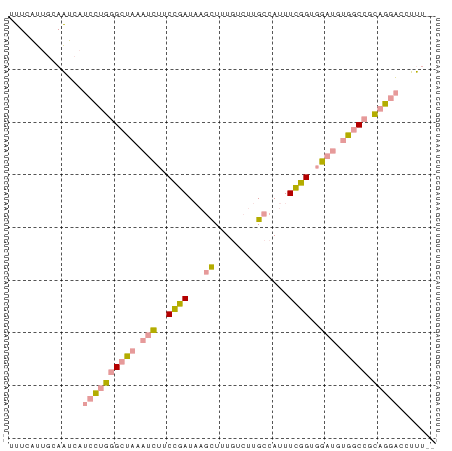
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:49:38 2011