| Sequence ID | dm3.chr3R |
|---|---|
| Location | 366,195 – 366,274 |
| Length | 79 |
| Max. P | 0.649030 |

| Location | 366,195 – 366,274 |
|---|---|
| Length | 79 |
| Sequences | 3 |
| Columns | 81 |
| Reading direction | forward |
| Mean pairwise identity | 60.17 |
| Shannon entropy | 0.55442 |
| G+C content | 0.45200 |
| Mean single sequence MFE | -17.14 |
| Consensus MFE | -9.46 |
| Energy contribution | -8.47 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.31 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.649030 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 366195 79 + 27905053 AGUGCCUUACACA--AUGGCUCUGCACGGACUGAACGUAGCAGAUUUCCAUUUAACGGUUACACUCUGGGUGACCAUCGUC .(((.....)))(--((((.((((((((.......))).)))))...)))))....((((((.(....)))))))...... ( -22.70, z-score = -1.64, R) >droSim1.chr3R 415646 74 + 27517382 -----AUUCCACA--AAGGCUCUGCACGGACUGAGCGUAGCAGAUUUCCAUAUAACGGUUACACUUUGGGUGCCCAUCGUC -----...((.((--(((((((..........))))(((((..((......))....))))).)))))))........... ( -15.40, z-score = 0.39, R) >droMoj3.scaffold_6540 14374638 69 + 34148556 AUUGCUUUGCAUUUCAAUGCUCUGCACCAAUCAAGAGUCGUAGGGU-----------AUUCUACUUUUAGUAACAAUCAU- .(((((.........(((((((((((((......).)).)))))))-----------)))........))))).......- ( -13.33, z-score = -1.57, R) >consensus A_UGCAUUACACA__AAGGCUCUGCACGGACUGAGCGUAGCAGAUUUCCAU_UAACGGUUACACUUUGGGUGACCAUCGUC .................((.((((((((.......))).))))).................(((.....))).))...... ( -9.46 = -8.47 + -0.99)

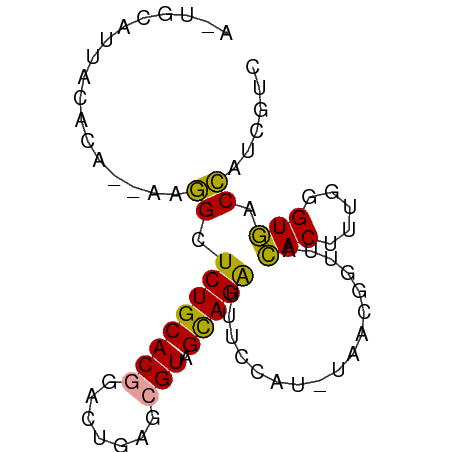

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:49:37 2011