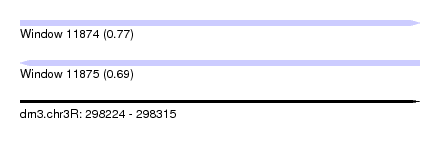
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 298,224 – 298,315 |
| Length | 91 |
| Max. P | 0.765655 |

| Location | 298,224 – 298,315 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 66.36 |
| Shannon entropy | 0.64224 |
| G+C content | 0.55056 |
| Mean single sequence MFE | -31.98 |
| Consensus MFE | -13.75 |
| Energy contribution | -13.48 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.61 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.765655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
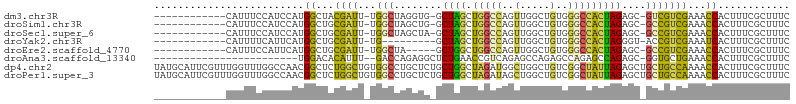
>dm3.chr3R 298224 91 + 27905053 ------------CAUUUCCAUCCAUGGCUACGAUU-UGGCUAGGUG-GCUAGCUGGCCAGUUGGCUGUGGGCCACUAGAGC-GUCGUCGAAACCACUUUCGCUUUC ------------.............(((.(((((.-..(((.((((-(((.((.((((....)))))).)))))))..)))-))))).((((....)))))))... ( -28.50, z-score = -0.33, R) >droSim1.chr3R 358623 91 + 27517382 ------------CAUUUCCAUCCAUGGCUGCGAUU-UGGCUAGCUG-GCUAGCUGGCCAGUUGGCUGUGGGCCACUAGAGC-GCCGUCGAAACCACUUUCGCUUUC ------------........((.(((((..(((((-.((((((((.-...)))))))))))))(((.(((....))).)))-))))).))................ ( -32.30, z-score = -0.81, R) >droSec1.super_6 404111 91 + 4358794 ------------CAUUUCCAUCCAUGGCUGCGAUU-UGGCUAGCUA-GCUAGCUGGCCAGUUGGCUGUGGGCCACUAGAGC-GCCGUCGAAACCACUUUCGCUUUC ------------........((.(((((..(((((-.((((((((.-...)))))))))))))(((.(((....))).)))-))))).))................ ( -32.60, z-score = -1.26, R) >droYak2.chr3R 677779 83 + 28832112 ------------CAUUUUCAUUCAUGGCUGCGAUU-UG---------GCUAGCUGGCCAGUUGGCUGUGGGCCACUAGGGU-ACCGUCGAAAUCACUUUCGCUUUC ------------........(((((((.(((..(.-((---------(((.((.((((....)))))).)))))..)..))-))))).)))............... ( -21.20, z-score = 0.33, R) >droEre2.scaffold_4770 645761 87 + 17746568 ------------CAUUUCCAUUCAUGGCUGCGAUU-UGGCUA-----GCUGGCUGGCCAGUUGGCUGUGGGCCACUAGAGC-GCCGUCGAAACCACUUUCGCUUUC ------------........((((((((..(((((-.(((((-----(....)))))))))))(((.(((....))).)))-))))).)))............... ( -29.30, z-score = -0.62, R) >droAna3.scaffold_13340 8021509 79 - 23697760 ------------------------UGGACACAUUU--GACCAGAGGCUCUGAACCGUCAGAGCCAGAGCCAGAGCCAGAGC-GGUGCUGAAACCACUUUCGCUUUC ------------------------(((.((....)--).)))..((((((((....))))))))......(((((.((((.-(((......))).)))).))))). ( -28.10, z-score = -2.43, R) >dp4.chr2 6321916 106 - 30794189 UAUGCAUUCGUUUGGUUUGGCCAACGGCUCUGGCUGUGGCCUGCUCUGCUGGCUAGAUGGCUGGCUGUCGGCUAUUAGAGCUGCUGCCAAAACCACUUUCGCUUUC ............(((((((((...((((((((...((((((.((...((..(((....)))..)).)).))))))))))))))..))).))))))........... ( -41.90, z-score = -2.30, R) >droPer1.super_3 122760 106 - 7375914 UAUGCAUUCGUUUGGUUUGGCCAACGGCUCUGGCUGUGGCCUGCUCUGCUGGCUAGAUAGCUGGCUGUCGGCUAUUAGAGCUGCUGCCAAAACCACUUUCGCUUUC ............(((((((((...((((((((...((((((.((...((..(((....)))..)).)).))))))))))))))..))).))))))........... ( -41.90, z-score = -2.45, R) >consensus ____________CAUUUCCAUCCAUGGCUGCGAUU_UGGCUAGCUG_GCUAGCUGGCCAGUUGGCUGUGGGCCACUAGAGC_GCCGUCGAAACCACUUUCGCUUUC .........................((...((((..(((((......(((((((....)))))))....)))))...........))))...))............ (-13.75 = -13.48 + -0.27)

| Location | 298,224 – 298,315 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 66.36 |
| Shannon entropy | 0.64224 |
| G+C content | 0.55056 |
| Mean single sequence MFE | -27.87 |
| Consensus MFE | -13.92 |
| Energy contribution | -13.27 |
| Covariance contribution | -0.65 |
| Combinations/Pair | 1.60 |
| Mean z-score | -0.79 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.690295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
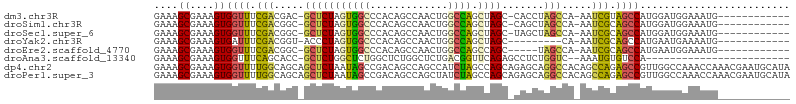
>dm3.chr3R 298224 91 - 27905053 GAAAGCGAAAGUGGUUUCGACGAC-GCUCUAGUGGCCCACAGCCAACUGGCCAGCUAGC-CACCUAGCCA-AAUCGUAGCCAUGGAUGGAAAUG------------ ....((....))((((.(((....-(((...(((((.....(((....)))......))-)))..)))..-..))).)))).............------------ ( -24.70, z-score = -0.22, R) >droSim1.chr3R 358623 91 - 27517382 GAAAGCGAAAGUGGUUUCGACGGC-GCUCUAGUGGCCCACAGCCAACUGGCCAGCUAGC-CAGCUAGCCA-AAUCGCAGCCAUGGAUGGAAAUG------------ ....((((...(((((.....(((-(((((((((((.....))).)))))..)))..))-)....)))))-..))))..(((....))).....------------ ( -28.40, z-score = -0.04, R) >droSec1.super_6 404111 91 - 4358794 GAAAGCGAAAGUGGUUUCGACGGC-GCUCUAGUGGCCCACAGCCAACUGGCCAGCUAGC-UAGCUAGCCA-AAUCGCAGCCAUGGAUGGAAAUG------------ ....((....))((((.(((.(((-((((((((((((...........))))).)))).-.)))..))).-..))).)))).............------------ ( -27.30, z-score = 0.28, R) >droYak2.chr3R 677779 83 - 28832112 GAAAGCGAAAGUGAUUUCGACGGU-ACCCUAGUGGCCCACAGCCAACUGGCCAGCUAGC---------CA-AAUCGCAGCCAUGAAUGAAAAUG------------ ....((....(((((((.(..(((-...((((((((.....))).)))))...)))..)---------.)-)))))).))..............------------ ( -19.30, z-score = -0.68, R) >droEre2.scaffold_4770 645761 87 - 17746568 GAAAGCGAAAGUGGUUUCGACGGC-GCUCUAGUGGCCCACAGCCAACUGGCCAGCCAGC-----UAGCCA-AAUCGCAGCCAUGAAUGGAAAUG------------ ....((((...(((((.....(((-...((((((((.....))).)))))...)))...-----.)))))-..))))..(((....))).....------------ ( -28.10, z-score = -0.97, R) >droAna3.scaffold_13340 8021509 79 + 23697760 GAAAGCGAAAGUGGUUUCAGCACC-GCUCUGGCUCUGGCUCUGGCUCUGACGGUUCAGAGCCUCUGGUC--AAAUGUGUCCA------------------------ ...(((.(.((((((......)))-))).).))).(((((..((((((((....))))))))...))))--)..........------------------------ ( -29.60, z-score = -1.79, R) >dp4.chr2 6321916 106 + 30794189 GAAAGCGAAAGUGGUUUUGGCAGCAGCUCUAAUAGCCGACAGCCAGCCAUCUAGCCAGCAGAGCAGGCCACAGCCAGAGCCGUUGGCCAAACCAAACGAAUGCAUA ....(((....((((((.((((((.(((((....(((....((..((......))..)).).)).(((....)))))))).))).)))))))))......)))... ( -32.10, z-score = -1.30, R) >droPer1.super_3 122760 106 + 7375914 GAAAGCGAAAGUGGUUUUGGCAGCAGCUCUAAUAGCCGACAGCCAGCUAUCUAGCCAGCAGAGCAGGCCACAGCCAGAGCCGUUGGCCAAACCAAACGAAUGCAUA ....(((....((((((.((((((.(((((....(((....((..(((....)))..)).).)).(((....)))))))).))).)))))))))......)))... ( -33.50, z-score = -1.57, R) >consensus GAAAGCGAAAGUGGUUUCGACGGC_GCUCUAGUGGCCCACAGCCAACUGGCCAGCCAGC_CAGCUAGCCA_AAUCGCAGCCAUGGAUGGAAAUG____________ ....((....))((((.(((.....(((((((((((.............)))).)))).......))).....))).))))......................... (-13.92 = -13.27 + -0.65)
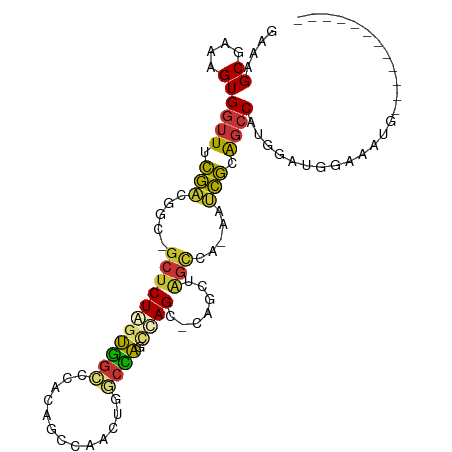
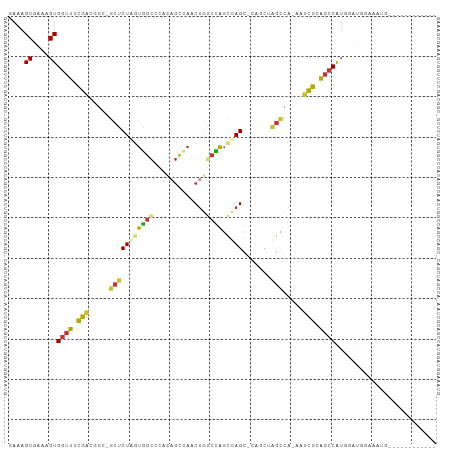
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:49:32 2011