| Sequence ID | dm3.chr3R |
|---|---|
| Location | 259,414 – 259,492 |
| Length | 78 |
| Max. P | 0.778867 |

| Location | 259,414 – 259,492 |
|---|---|
| Length | 78 |
| Sequences | 15 |
| Columns | 83 |
| Reading direction | forward |
| Mean pairwise identity | 71.41 |
| Shannon entropy | 0.63778 |
| G+C content | 0.45400 |
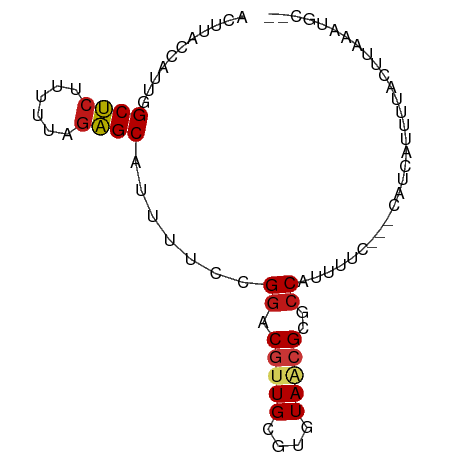
| Mean single sequence MFE | -16.42 |
| Consensus MFE | -10.52 |
| Energy contribution | -10.37 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.18 |
| Mean z-score | -0.40 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.778867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
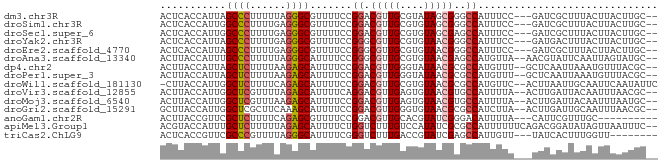
>dm3.chr3R 259414 78 + 27905053 ACUCACCAUUAGCCCUUUUUAGGGCGUUUUCCGGACGUUGCGUAUAGCGGGCCAUUUCC---GAUCGCUUUACUUACUUGC-- ...........(((((....)))))(((.....))).....(((.(((((..(......---).))))).)))........-- ( -17.30, z-score = -0.14, R) >droSim1.chr3R 319127 78 + 27517382 ACUCACCAUUGGCCCUUUUGAGGGCGUUUUCCGGACGUUGCGUGUAGCGGGCCAUUUCC---GAUCGCUUUACUUACUUGC-- .........((((((.......((((((.....))))))((.....)))))))).....---...................-- ( -18.80, z-score = 0.48, R) >droSec1.super_6 364905 78 + 4358794 ACUCACCAUUGGCCCUUUUGAGGGCGUUUUCCGGACGUUGCGUGUAGCGAGCCAUUUCC---GAUCGCUUUACUUACUUGC-- ...........(((((....)))))(((.....))).....(((.(((((..(......---).))))).)))........-- ( -17.20, z-score = 0.44, R) >droYak2.chr3R 635209 78 + 28832112 ACUCACCAUUAGCCCUUUUGAGGGCGUUUUCCGGGCGUUGCGUGUAACGGGCCAUUUCC---GAUGACUUUACUUACUUGC-- ...........(((((....)))))(((...((((.((.((.((...)).)).)).)))---)..))).............-- ( -17.50, z-score = 0.44, R) >droEre2.scaffold_4770 605508 78 + 17746568 ACUCACCAUUAGCCCUUUUGAGGGCGUUUUCCGGGCGUUGCGUGUAACGGGCCAUUUCC---GAUCGCUUUACUUACUUGC-- ...........(((((....)))))......((((.((.((.((...)).)).)).)))---)..................-- ( -16.20, z-score = 1.16, R) >droAna3.scaffold_13340 11256080 79 + 23697760 ACUUACCAUUUGCCCUUUUUAGGGCAUUUUCCGGGCGUUGCGUGUAACGAGCCAUGUUA--AACGUAUUCAAUUAGUAUGC-- .....((...((((((....))))))......))((((((((((........)))))..--)))))...............-- ( -18.20, z-score = -0.60, R) >dp4.chr2 6278680 79 - 30794189 ACUUACCAUUAGCUCUUUUAAGAGCAUUUUCCGGACGUUGGGUAUAACGCGCCAUGUUU--GCUCAAUUAAAUGUUUACGC-- ......((((.(((((....)))))...........((((((((.((((.....)))))--)))))))..)))).......-- ( -15.00, z-score = -0.34, R) >droPer1.super_3 78961 79 - 7375914 ACUUACCAUUAGCUCUUUUAAGAGCAUUUUCCGGACGUUGGGUAUAACGCGCCAUGUUU--GCUCAAUUAAAUGUUUACGC-- ......((((.(((((....)))))...........((((((((.((((.....)))))--)))))))..)))).......-- ( -15.00, z-score = -0.34, R) >droWil1.scaffold_181130 13698984 80 - 16660200 -CUUACCAUUGGCUCUUUUCAGAGCAUUUUCCGGACGUUGCGUGUAACGCGCCAUGUUC--ACUUAAUUGCAAUUCAAUAUUC -......(((((((((....))))).......((((((.(((((...))))).))))))--..............)))).... ( -20.30, z-score = -2.82, R) >droVir3.scaffold_12855 7376905 79 - 10161210 ACUUACCAUUGGCUCGUUUUAGAGCAUUUUCAGGACGUUGAGUGUAACGUGCCAUUUUA--ACUUGAUUACAAUUUAACGC-- .......((((((((......)))).......((((((((....)))))).))......--.........)))).......-- ( -14.80, z-score = -0.72, R) >droMoj3.scaffold_6540 31269002 79 + 34148556 ACUUACCAUUGGCUCGUUUAAGAGCAUUUUCCGGACGUUGAGUGUAACGUGCCAUUUUA--ACUUGAUUACAAUUUAAUGC-- ......(((((((((......)))).......((((((((....)))))).))......--..............))))).-- ( -15.40, z-score = -0.88, R) >droGri2.scaffold_15291 2898 79 + 6112 GCUUACCAUUGGCUCGCUUCAAAGCAUUUUCCGGACGUUGGGUGUAACGCGCCAUCUUA--ACUUGAUUGCAAUUUAACGC-- (((.......)))..(((....)))..........(((((((((.....))))(((...--....))).......))))).-- ( -12.10, z-score = 1.68, R) >anoGam1.chr2R 42441310 70 - 62725911 ACUUACCGUUCGCUCUUUUCAGAGCGUUUUCCGGACGUUGCACGUAUCGGGACAUUUUA---CAUUCGUUUGC---------- ..........((((((....))))))..(((((((((.....))).)))))).......---...........---------- ( -16.20, z-score = -1.27, R) >apiMel3.Group1 11407487 81 + 25854376 ACGUACCAUUUGCUCUUUUUAGAGCAUUUUCUGGUCUUUGUCCAUAUCGCGCCAUUUUUUCAGACGGAUAUAGUUAAUUUC-- ....((((..((((((....)))))).....))))((.(((((...((..............)).))))).))........-- ( -14.04, z-score = -1.56, R) >triCas2.ChLG9 12803226 72 - 15222296 ACUCACCGUUCGCCCGUUUUAGGGCAUUUUCGGGUCUUUGACCGUAUCGAGCCAUUGUU---UAUCACUUUGGUU-------- .(((.......((((......)))).......((((...)))).....)))(((..((.---....))..)))..-------- ( -18.30, z-score = -1.58, R) >consensus ACUUACCAUUGGCUCUUUUUAGAGCAUUUUCCGGACGUUGCGUGUAACGCGCCAUUUUC___CAUCAUUUUACUUAAAUGC__ ...........((((......)))).......((.(((((....)))))..)).............................. (-10.52 = -10.37 + -0.15)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:49:29 2011