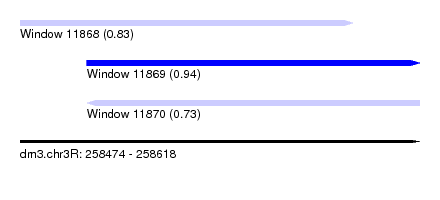
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 258,474 – 258,618 |
| Length | 144 |
| Max. P | 0.936037 |

| Location | 258,474 – 258,594 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.51 |
| Shannon entropy | 0.44649 |
| G+C content | 0.51889 |
| Mean single sequence MFE | -46.37 |
| Consensus MFE | -21.92 |
| Energy contribution | -20.24 |
| Covariance contribution | -1.68 |
| Combinations/Pair | 1.70 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.829066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 258474 120 + 27905053 GCGCUGCAGCGUGGAACAACUGGUUGCCGGCCUUGCUAAAGACCAUGGCUAAUUUUUGGUAGUAGUUGGCCAUGGUUUUGGGCACGGGUGUCUUCUUGGACAGAGCCAUCAGGCCAUGGA .........(((((.....(((((.(((.((((((((.((((((((((((((((.........)))))))))))))))).)))).))))((((....)))).).)).))))).))))).. ( -49.60, z-score = -2.13, R) >droSim1.chr3R 318185 120 + 27517382 GCGCUGCAGCGUGAAACAACUGGUUGCCGGCCUUGCUGAAAACCAUGGCUAAUUUUUGGUAGUAGUUGGCCAUGGUUUUGGGCACAGGUGUCUUCUUGGACAGAGCCAUCAGGCCAUGGA ((((....)))).......(((((.(((.((((((((.((((((((((((((((.........)))))))))))))))).)))).))))((((....)))).).)).)))))........ ( -50.40, z-score = -2.61, R) >droSec1.super_6 363963 120 + 4358794 GCGCUGCAGCGUGAAACAACUGGUUGCCGGCCUUGCUGAAAACCAUGGCUAAUUUUUGGUAGUAGUUGGCCAUGGUUUUGGGCACAGGUGUCUUCUUGGACAGAGCCAUCAGGCCAUGGA ((((....)))).......(((((.(((.((((((((.((((((((((((((((.........)))))))))))))))).)))).))))((((....)))).).)).)))))........ ( -50.40, z-score = -2.61, R) >droYak2.chr3R 634267 120 + 28832112 GGGCUGCAGCGUGGAACAACUGGUUGCCAGCCUUGCUAAAGACCAUGGCUAAUUUUUGGUAAUAGUUGGCCAUUGUUUUGGGUACGGGUGUCUUUUUGGAUAAAGCCAUCAAGCCGUGGA .(((((((((..(......)..)))).)))))(..(..(((((.((((((((((.........)))))))))).))))).(((...((((.((((......)))).))))..))))..). ( -38.10, z-score = -0.26, R) >droEre2.scaffold_4770 604568 120 + 17746568 GGGCUGCAGCGUGGAACAACUGGUUGCCAGCCUUGCUGAAGACCAUGGCCAAUUUUUGGUAAUAGUUGGCCAUGGUUUUGGGCACGGGUGUCUUCUUGGACAGAGCCAUCAGGCCAUGGA .........(((((.....(((((.(((.((((((((.((((((((((((((((.........)))))))))))))))).)))).))))((((....)))).).)).))))).))))).. ( -53.90, z-score = -3.09, R) >droAna3.scaffold_13340 11255127 120 + 23697760 GAGCGGCAGCGUGGAACAACUGGUUGCCAGCCUUGCUGAAGACCAUCGCCAGUUUCUGAUAGUAAUUGGCCAUGGUUUUGGGCACCGGCGUCUUCUUGGACAGGGCCAUAAGACCGUGAA ....((((((..(......)..)))))).(((.((((.((((((((.(((((((.(.....).))))))).)))))))).))))..)))(((((..(((......))).)))))...... ( -47.60, z-score = -2.03, R) >dp4.chr2 6277710 120 - 30794189 GGGCGGCGGCAUGGAAAAGCUGAUUGCCUGCCUUGCUGAAGACCAUGGCAAGCUUCUGGUAAUAGUUCGCCAUCGUUUUGGGCACGGGCGUCUUCUUGGAGAGGGCCAUCAGGCCGUGGA ...(.(((((.(....)..(((((.((((((((((((.(((((.(((((.((((.........)))).))))).))))).)))).)))).((((....)))))))).)))))))))).). ( -51.70, z-score = -1.78, R) >droPer1.super_3 77993 120 - 7375914 GGGCGGCGGCAUGGAAAAGCUGAUUGCCUGCCUUGCUGAAGACCAUGGCAAGCUUCUGGUAAUAGUUCGCCAUCGUUUUGGGCACGGGCGUCUUCUUGGAGAGGGCCAUCAGGCCGUGGA ...(.(((((.(....)..(((((.((((((((((((.(((((.(((((.((((.........)))).))))).))))).)))).)))).((((....)))))))).)))))))))).). ( -51.70, z-score = -1.78, R) >droWil1.scaffold_181130 13697995 120 - 16660200 AAGCAGCUGCAUGGAACAGUUGGUUGCCAGCCUUGCUGAAGACCAUUGCAAGUUUUUGGUAAUAGUUUGCCAUGGUCUUGGGCACGGGCGUCUUCUUGGAGAGAGCCAUCAGACCGUGAA .........(((((......(((((....((((((((.((((((((.((((..(.........)..)))).)))))))).)))).)))).((((....)))).))))).....))))).. ( -42.70, z-score = -1.18, R) >droVir3.scaffold_12855 7375934 120 - 10161210 AAGCGGCUGCAUGGAAUAGCUGGUUACCAGCCUUGCUAAAUACCAUAGCCAGCUUCUGAUAGUAAUUGGCCAUGGUUUUGGGCACCGGCGUCUUCUUGGACAUGGCCAUCAGGCCGUGAA ..((((((..((((....(((((...)))))..((((....(((((.(((((((......)))...)))).)))))....))))(((..((((....)))).)))))))..))))))... ( -44.70, z-score = -1.35, R) >droMoj3.scaffold_6540 31268019 120 + 34148556 AAGCGGCUGCAUGGAAGAGCUGAUUGCCAGCCUUGCUGAAGACCAUAGCCAAUUUCUGAUAAUAAUUAGCCAUGGUUUUUGGCACCGGCGUCUUCUUGGACAUGGCCAUCAGACCGUGAA .........(((((.....(((((.(((((((.(((..((((((((.((.((((.........)))).)).))))))))..)))..)))((((....)))).)))).))))).))))).. ( -46.50, z-score = -3.39, R) >droGri2.scaffold_15291 1900 120 + 6112 AAGCGGCCGCAUGGAAGAGCUGAUUGCCGGCCUUGCUGAAAACCAUGGCAAGUUUCUGAUAAUAAUUUGCCAUGGUUUUGGGCACCGGCGUCUUCUUGGACAUGGCCAUUAGGCCGUGAA ..((((((..((((...........(((((...((((.((((((((((((((((.........)))))))))))))))).)))))))))((((....))))....))))..))))))... ( -59.40, z-score = -5.68, R) >anoGam1.chr2R 42440303 120 - 62725911 GUGCCGACGCGUGGAACAGCUGAUUGCCCGCCUUCCAGAACACCAUCGCCAGCUUGGUGUAGUACAGGGCCAUCGUUUUCGGCAGCGGCGUCUUCUUGCUCAGCUGCAUCAGCCCGUGAA .......((((.((....(((((..((..(((((.....((((((.........)))))).....)))))....))..))))).(((((.............))))).....)))))).. ( -37.02, z-score = 1.25, R) >apiMel3.Group1 11405980 120 + 25854376 AUGCAGCAGCAUGGAAAAGAUAGUUUCCUGCUUUCCAGAAAACCAUAGCCAAUUUUUGGUAGUAAUUGGCCAUGGUUUUUGGUACUGGAAGUUUCUUAGAUAAAUUCAUCAUACCAUGUA ........((((((..((((...(((((......(((((((.((((.(((((((.........))))))).)))))))))))....)))))..)))).(((......)))...)))))). ( -35.90, z-score = -3.11, R) >triCas2.ChLG9 12801850 120 - 15222296 GAGCUGCCGCGUGGAAGAGGUAAUUUCCGGCCUUCCAGAAAACCAUGGCCAGUUUUUGGUAGUAAUUGGCCAUUGUUUUAGGCACCGGCAUUUUUUUAGACAUGUUCAUUAGGUUAUGGA ....(((((.(((((((......))))).((((.....(((((.((((((((((.........)))))))))).))))))))))))))))..........((((..(....)..)))).. ( -36.00, z-score = -1.31, R) >consensus GAGCGGCAGCGUGGAACAGCUGGUUGCCAGCCUUGCUGAAGACCAUGGCCAGUUUUUGGUAGUAGUUGGCCAUGGUUUUGGGCACCGGCGUCUUCUUGGACAGAGCCAUCAGGCCGUGGA .........(((((.....(((((((((.(((......(((((.((.(((((((.........))))))).)).))))).)))...)))).......((......))))))).))))).. (-21.92 = -20.24 + -1.68)
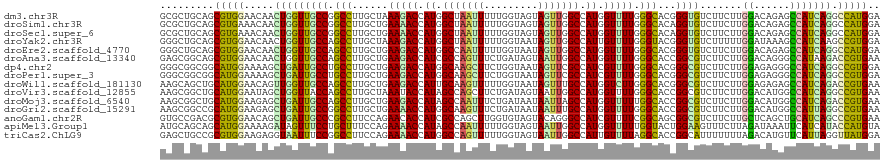
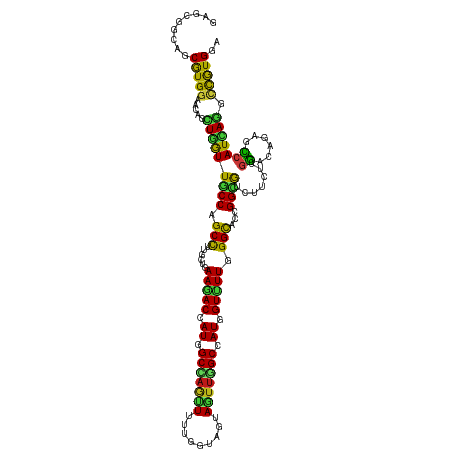
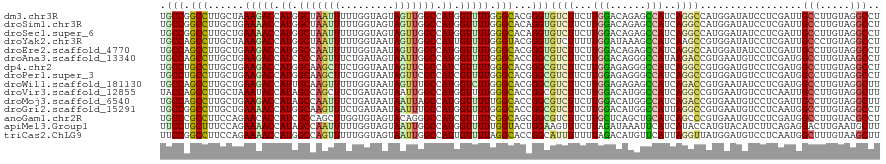
| Location | 258,498 – 258,618 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.52 |
| Shannon entropy | 0.39927 |
| G+C content | 0.50500 |
| Mean single sequence MFE | -46.91 |
| Consensus MFE | -20.06 |
| Energy contribution | -19.45 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.57 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.936037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
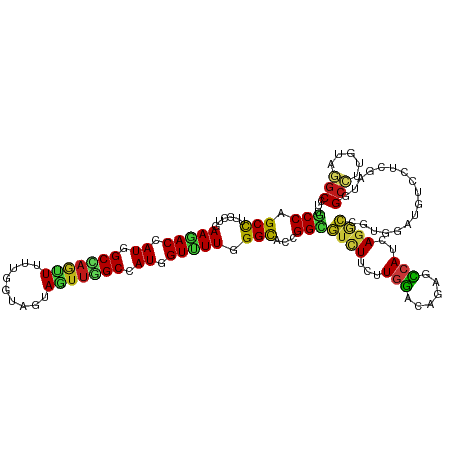
>dm3.chr3R 258498 120 + 27905053 UGCCGGCCUUGCUAAAGACCAUGGCUAAUUUUUGGUAGUAGUUGGCCAUGGUUUUGGGCACGGGUGUCUUCUUGGACAGAGCCAUCAGGCCAUGGAUAUCCUCGAUUGCCUUGUAGGCCU ....(((((.((..((((((((((((((((.........))))))))))))))))(((((.((((((((...(((...((....))...))).)))))))).....))))).))))))). ( -49.90, z-score = -2.69, R) >droSim1.chr3R 318209 120 + 27517382 UGCCGGCCUUGCUGAAAACCAUGGCUAAUUUUUGGUAGUAGUUGGCCAUGGUUUUGGGCACAGGUGUCUUCUUGGACAGAGCCAUCAGGCCAUGGAUAUCCUCGAUUGCCUUGUAGGCCU ....(((((.(((.((((((((((((((((.........)))))))))))))))).)))(((((.((..((..(((((..(((....)))..))....)))..))..)))))))))))). ( -52.60, z-score = -3.60, R) >droSec1.super_6 363987 120 + 4358794 UGCCGGCCUUGCUGAAAACCAUGGCUAAUUUUUGGUAGUAGUUGGCCAUGGUUUUGGGCACAGGUGUCUUCUUGGACAGAGCCAUCAGGCCAUGGAUAUCCUCGAUUGCCUUGUAGGCCU ....(((((.(((.((((((((((((((((.........)))))))))))))))).)))(((((.((..((..(((((..(((....)))..))....)))..))..)))))))))))). ( -52.60, z-score = -3.60, R) >droYak2.chr3R 634291 120 + 28832112 UGCCAGCCUUGCUAAAGACCAUGGCUAAUUUUUGGUAAUAGUUGGCCAUUGUUUUGGGUACGGGUGUCUUUUUGGAUAAAGCCAUCAAGCCGUGGAUAUCCUCGAUUGCCCUGUAGGCCU .....((((.(((.(((((.((((((((((.........)))))))))).))))).)))(((((.((..((..(((((...((((......)))).)))))..))..))))))))))).. ( -39.90, z-score = -1.51, R) >droEre2.scaffold_4770 604592 120 + 17746568 UGCCAGCCUUGCUGAAGACCAUGGCCAAUUUUUGGUAAUAGUUGGCCAUGGUUUUGGGCACGGGUGUCUUCUUGGACAGAGCCAUCAGGCCAUGGAUAUCCUCGAUUGCCUUGUAGGCCU .....((((((((.((((((((((((((((.........)))))))))))))))).)))).))))((((....))))((.(((((.((((.((.((.....)).)).)))).)).))))) ( -52.80, z-score = -3.43, R) >droAna3.scaffold_13340 11255151 120 + 23697760 UGCCAGCCUUGCUGAAGACCAUCGCCAGUUUCUGAUAGUAAUUGGCCAUGGUUUUGGGCACCGGCGUCUUCUUGGACAGGGCCAUAAGACCGUGAAUGUCCUCGAUGGCCUUGUAAGCCU .((..(((.((((.((((((((.(((((((.(.....).))))))).)))))))).))))..)))..........((((((((((..(((.......)))....))))))))))..)).. ( -49.00, z-score = -3.16, R) >dp4.chr2 6277734 120 - 30794189 UGCCUGCCUUGCUGAAGACCAUGGCAAGCUUCUGGUAAUAGUUCGCCAUCGUUUUGGGCACGGGCGUCUUCUUGGAGAGGGCCAUCAGGCCGUGGAUGUCCUCGAUGGCCUUGUAGGCCU .(((((...((((.(((((.(((((.((((.........)))).))))).))))).))))))))).((((....))))(((((((.(((((((.((.....)).))))))).)).))))) ( -53.80, z-score = -2.56, R) >droPer1.super_3 78017 120 - 7375914 UGCCUGCCUUGCUGAAGACCAUGGCAAGCUUCUGGUAAUAGUUCGCCAUCGUUUUGGGCACGGGCGUCUUCUUGGAGAGGGCCAUCAGGCCGUGGAUGUCCUCGAUGGCCUUGUAGGCCU .(((((...((((.(((((.(((((.((((.........)))).))))).))))).))))))))).((((....))))(((((((.(((((((.((.....)).))))))).)).))))) ( -53.80, z-score = -2.56, R) >droWil1.scaffold_181130 13698019 120 - 16660200 UGCCAGCCUUGCUGAAGACCAUUGCAAGUUUUUGGUAAUAGUUUGCCAUGGUCUUGGGCACGGGCGUCUUCUUGGAGAGAGCCAUCAGACCGUGAAUAUCCUCGAUGGCCUUGUAGGCUU .(((.((((((((.((((((((.((((..(.........)..)))).)))))))).)))).))))...........(((.((((((((............)).)))))))))...))).. ( -45.50, z-score = -2.33, R) >droVir3.scaffold_12855 7375958 120 - 10161210 UACCAGCCUUGCUAAAUACCAUAGCCAGCUUCUGAUAGUAAUUGGCCAUGGUUUUGGGCACCGGCGUCUUCUUGGACAUGGCCAUCAGGCCGUGAAUGUCCUCAAUUGCCUUGUAGGCUU ....(((((((((....(((((.(((((((......)))...)))).)))))....))))..((((.......((((((((((....))))....)))))).....))))....))))). ( -43.60, z-score = -2.20, R) >droMoj3.scaffold_6540 31268043 120 + 34148556 UGCCAGCCUUGCUGAAGACCAUAGCCAAUUUCUGAUAAUAAUUAGCCAUGGUUUUUGGCACCGGCGUCUUCUUGGACAUGGCCAUCAGACCGUGAAUGUCCUCGAUUGCCUUGUAGGCUU ....((((((((..((((((((.((.((((.........)))).)).))))))))..)))..((((...((..((((((...(((......))).))))))..)).))))....))))). ( -41.00, z-score = -2.78, R) >droGri2.scaffold_15291 1924 120 + 6112 UGCCGGCCUUGCUGAAAACCAUGGCAAGUUUCUGAUAAUAAUUUGCCAUGGUUUUGGGCACCGGCGUCUUCUUGGACAUGGCCAUUAGGCCGUGAAUGUCCUCAAUGGCCUUGUAGGCCU .(((((...((((.((((((((((((((((.........)))))))))))))))).)))))))))((((....))))..((((((.(((((((((......)).))))))).)).)))). ( -62.70, z-score = -7.10, R) >anoGam1.chr2R 42440327 120 - 62725911 UGCCCGCCUUCCAGAACACCAUCGCCAGCUUGGUGUAGUACAGGGCCAUCGUUUUCGGCAGCGGCGUCUUCUUGCUCAGCUGCAUCAGCCCGUGAAUGUCCUCGAUGGCCUUGUACGCCU .....((........((((((.........)))))).((((((((((((((....(((((((((((......))))..))))).(((.....))).))....)))))))))))))))).. ( -46.70, z-score = -2.61, R) >apiMel3.Group1 11406004 120 + 25854376 UUCCUGCUUUCCAGAAAACCAUAGCCAAUUUUUGGUAGUAAUUGGCCAUGGUUUUUGGUACUGGAAGUUUCUUAGAUAAAUUCAUCAUACCAUGUACAUCUUCAGAGGACUUGAAUGCUU .((((.((.....(((((((((.(((((((.........))))))).))))))))).((((((((((...))).(((......)))...))).))))......))))))........... ( -28.50, z-score = -1.11, R) >triCas2.ChLG9 12801874 120 - 15222296 UUCCGGCCUUCCAGAAAACCAUGGCCAGUUUUUGGUAGUAAUUGGCCAUUGUUUUAGGCACCGGCAUUUUUUUAGACAUGUUCAUUAGGUUAUGGAUGUCCUCAAUGGCUUUGUAAGCUU ..((((....((..(((((.((((((((((.........)))))))))).))))).))..))))..........((((..(((((......)))))))))......((((.....)))). ( -31.20, z-score = -0.54, R) >consensus UGCCAGCCUUGCUGAAGACCAUGGCCAGUUUUUGGUAGUAGUUGGCCAUGGUUUUGGGCACCGGCGUCUUCUUGGACAGAGCCAUCAGGCCGUGGAUGUCCUCGAUGGCCUUGUAGGCCU .(((.(((......(((((.((.(((((((.........))))))).)).))))).)))...)))((((...(((......)))..)))).................(((.....))).. (-20.06 = -19.45 + -0.61)
| Location | 258,498 – 258,618 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.52 |
| Shannon entropy | 0.39927 |
| G+C content | 0.50500 |
| Mean single sequence MFE | -40.18 |
| Consensus MFE | -20.46 |
| Energy contribution | -20.38 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.64 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.725210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
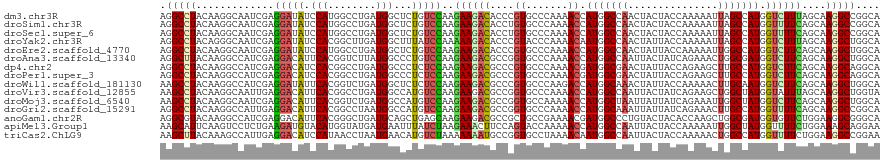
>dm3.chr3R 258498 120 - 27905053 AGGCCUACAAGGCAAUCGAGGAUAUCCAUGGCCUGAUGGCUCUGUCCAAGAAGACACCCGUGCCCAAAACCAUGGCCAACUACUACCAAAAAUUAGCCAUGGUCUUUAGCAAGGCCGGCA .(((((....((((..((.((........((((....)))).((((......))))))))))))....((((((((...................))))))))........))))).... ( -37.21, z-score = -1.19, R) >droSim1.chr3R 318209 120 - 27517382 AGGCCUACAAGGCAAUCGAGGAUAUCCAUGGCCUGAUGGCUCUGUCCAAGAAGACACCUGUGCCCAAAACCAUGGCCAACUACUACCAAAAAUUAGCCAUGGUUUUCAGCAAGGCCGGCA .(((((...(((...((..(((((.....((((....)))).)))))..)).....))).(((..(((((((((((...................)))))))))))..)))))))).... ( -39.91, z-score = -1.85, R) >droSec1.super_6 363987 120 - 4358794 AGGCCUACAAGGCAAUCGAGGAUAUCCAUGGCCUGAUGGCUCUGUCCAAGAAGACACCUGUGCCCAAAACCAUGGCCAACUACUACCAAAAAUUAGCCAUGGUUUUCAGCAAGGCCGGCA .(((((...(((...((..(((((.....((((....)))).)))))..)).....))).(((..(((((((((((...................)))))))))))..)))))))).... ( -39.91, z-score = -1.85, R) >droYak2.chr3R 634291 120 - 28832112 AGGCCUACAGGGCAAUCGAGGAUAUCCACGGCUUGAUGGCUUUAUCCAAAAAGACACCCGUACCCAAAACAAUGGCCAACUAUUACCAAAAAUUAGCCAUGGUCUUUAGCAAGGCUGGCA .((((....(((.......((....))(((((((..(((......)))..)))....)))).)))........))))..................((((..(((((....))))))))). ( -28.00, z-score = 0.52, R) >droEre2.scaffold_4770 604592 120 - 17746568 AGGCCUACAAGGCAAUCGAGGAUAUCCAUGGCCUGAUGGCUCUGUCCAAGAAGACACCCGUGCCCAAAACCAUGGCCAACUAUUACCAAAAAUUGGCCAUGGUCUUCAGCAAGGCUGGCA .(((((....((((..((.((........((((....)))).((((......))))))))))))....(((((((((((.............)))))))))))........))))).... ( -40.42, z-score = -1.42, R) >droAna3.scaffold_13340 11255151 120 - 23697760 AGGCUUACAAGGCCAUCGAGGACAUUCACGGUCUUAUGGCCCUGUCCAAGAAGACGCCGGUGCCCAAAACCAUGGCCAAUUACUAUCAGAAACUGGCGAUGGUCUUCAGCAAGGCUGGCA .((((.....)))).((..(((((.....((((....)))).)))))..))....((((((((..((.(((((.((((.((.(.....).)).)))).))))).))..))...)))))). ( -41.80, z-score = -1.58, R) >dp4.chr2 6277734 120 + 30794189 AGGCCUACAAGGCCAUCGAGGACAUCCACGGCCUGAUGGCCCUCUCCAAGAAGACGCCCGUGCCCAAAACGAUGGCGAACUAUUACCAGAAGCUUGCCAUGGUCUUCAGCAAGGCAGGCA ..((((....(((((((.(((.(......).))))))))))..............(((..(((..((.((.(((((((.((.........)).))))))).)).))..))).))))))). ( -42.40, z-score = -1.58, R) >droPer1.super_3 78017 120 + 7375914 AGGCCUACAAGGCCAUCGAGGACAUCCACGGCCUGAUGGCCCUCUCCAAGAAGACGCCCGUGCCCAAAACGAUGGCGAACUAUUACCAGAAGCUUGCCAUGGUCUUCAGCAAGGCAGGCA ..((((....(((((((.(((.(......).))))))))))..............(((..(((..((.((.(((((((.((.........)).))))))).)).))..))).))))))). ( -42.40, z-score = -1.58, R) >droWil1.scaffold_181130 13698019 120 + 16660200 AAGCCUACAAGGCCAUCGAGGAUAUUCACGGUCUGAUGGCUCUCUCCAAGAAGACGCCCGUGCCCAAGACCAUGGCAAACUAUUACCAAAAACUUGCAAUGGUCUUCAGCAAGGCUGGCA ..(((.....(((((((..((((.......)))))))))))..............(((..(((..((((((((.((((...............)))).))))))))..))).))).))). ( -39.16, z-score = -2.17, R) >droVir3.scaffold_12855 7375958 120 + 10161210 AAGCCUACAAGGCAAUUGAGGACAUUCACGGCCUGAUGGCCAUGUCCAAGAAGACGCCGGUGCCCAAAACCAUGGCCAAUUACUAUCAGAAGCUGGCUAUGGUAUUUAGCAAGGCUGGUA ..(((.....)))......((((((....((((....))))))))))........(((..(((..((.((((((((((.((.(.....).)).)))))))))).))..))).)))..... ( -44.30, z-score = -2.53, R) >droMoj3.scaffold_6540 31268043 120 - 34148556 AAGCCUACAAGGCAAUCGAGGACAUUCACGGUCUGAUGGCCAUGUCCAAGAAGACGCCGGUGCCAAAAACCAUGGCUAAUUAUUAUCAGAAAUUGGCUAUGGUCUUCAGCAAGGCUGGCA ..(((.....)))..((..((((((....((((....))))))))))..))....((((((((..((.(((((((((((((.........))))))))))))).))..))...)))))). ( -45.50, z-score = -3.67, R) >droGri2.scaffold_15291 1924 120 - 6112 AGGCCUACAAGGCCAUUGAGGACAUUCACGGCCUAAUGGCCAUGUCCAAGAAGACGCCGGUGCCCAAAACCAUGGCAAAUUAUUAUCAGAAACUUGCCAUGGUUUUCAGCAAGGCCGGCA .((((.....)))).....((((((....((((....))))))))))........((((((((..(((((((((((((...............)))))))))))))..))...)))))). ( -54.76, z-score = -5.92, R) >anoGam1.chr2R 42440327 120 + 62725911 AGGCGUACAAGGCCAUCGAGGACAUUCACGGGCUGAUGCAGCUGAGCAAGAAGACGCCGCUGCCGAAAACGAUGGCCCUGUACUACACCAAGCUGGCGAUGGUGUUCUGGAAGGCGGGCA ..(((((((.((((((((((..(......)..))...(((((.(.((........))))))))......)))))))).))))).((((((.(....)..)))))).(((.....))))). ( -41.30, z-score = 0.12, R) >apiMel3.Group1 11406004 120 - 25854376 AAGCAUUCAAGUCCUCUGAAGAUGUACAUGGUAUGAUGAAUUUAUCUAAGAAACUUCCAGUACCAAAAACCAUGGCCAAUUACUACCAAAAAUUGGCUAUGGUUUUCUGGAAAGCAGGAA ...........((((((..(((((..(((......)))....))))).......((((((.....((((((((((((((((.........)))))))))))))))))))))))).)))). ( -34.50, z-score = -3.34, R) >triCas2.ChLG9 12801874 120 + 15222296 AAGCUUACAAAGCCAUUGAGGACAUCCAUAACCUAAUGAACAUGUCUAAAAAAAUGCCGGUGCCUAAAACAAUGGCCAAUUACUACCAAAAACUGGCCAUGGUUUUCUGGAAGGCCGGAA ..(((.....)))((((..((((((.(((......)))...)))))).....))))(((((.((.(((((.(((((((...............))))))).)))))..))...))))).. ( -31.16, z-score = -1.72, R) >consensus AGGCCUACAAGGCCAUCGAGGACAUCCACGGCCUGAUGGCCCUGUCCAAGAAGACGCCCGUGCCCAAAACCAUGGCCAACUACUACCAAAAACUGGCCAUGGUCUUCAGCAAGGCUGGCA .(((((.............(((((.(((........)))...)))))..((((((....((.......)).(((((((...............))))))).))))))....))))).... (-20.46 = -20.38 + -0.08)
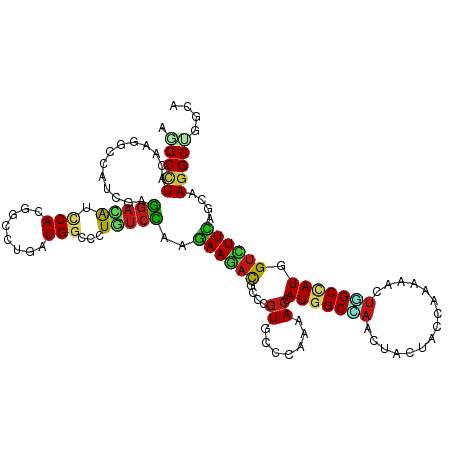
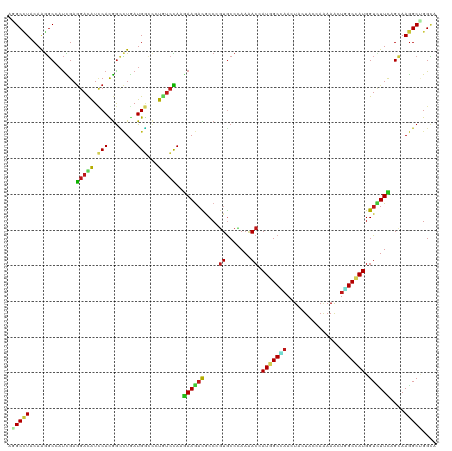
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:49:28 2011