
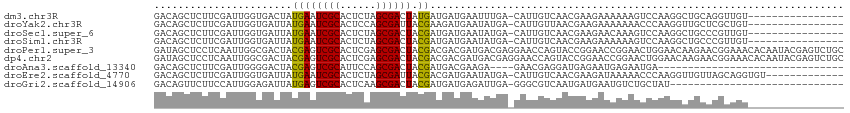
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 252,054 – 252,152 |
| Length | 98 |
| Max. P | 0.500000 |

| Location | 252,054 – 252,152 |
|---|---|
| Length | 98 |
| Sequences | 9 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 63.99 |
| Shannon entropy | 0.69694 |
| G+C content | 0.46398 |
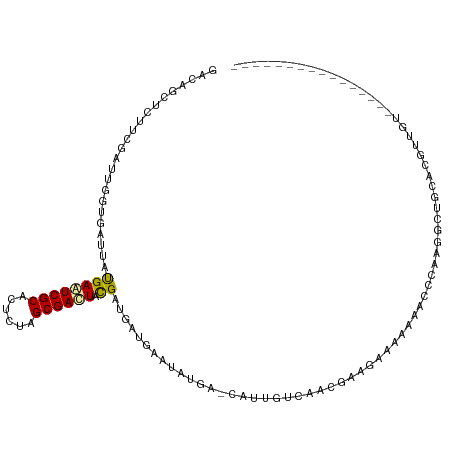
| Mean single sequence MFE | -26.01 |
| Consensus MFE | -7.79 |
| Energy contribution | -7.61 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr3R 252054 98 - 27905053 GACAGCUCUUCGAUUGGUGACUAUGAAUCGCACUCUAGCGACUAUGAUGAUGAAUUUGA-CAUUGUCAACGAAGAAAAAAGUCCAAGGCUGCAGGUUGU---------------- .(((((((((((...(....)......((((......))))...(((..(((.......-)))..))).))))))....(((.....)))....)))))---------------- ( -23.90, z-score = -0.63, R) >droYak2.chr3R 627077 98 - 28832112 GACAGCUCUUCGAUUGGUGAUUAUGAAUCGCACUCCAGCGAUUACGAAGAUGAAUAUGA-CAUUGUUAACGAAGAAAAAAACCCAAGGUUGCUCGCUGU---------------- .((((((((((((((.((..((...((((((......))))))...)).)).))).(((-(...)))).))))))....((((...))))....)))))---------------- ( -22.90, z-score = -1.31, R) >droSec1.super_6 356043 98 - 4358794 GACAGCUCUUCGAUUGGUGAUUAUGAAUCGCACUCUAGCGACUACGAUGAUGAAUAUGA-CAUUGUCAACGAAGAACAAAGUCCAAGGCUGCCCGUUGU---------------- .(((((((((((...............((((......))))....((..(((.......-)))..))..))))))....(((.....)))....)))))---------------- ( -22.90, z-score = -0.45, R) >droSim1.chr3R 311416 98 - 27517382 GACAGCUCUUCGAUUGGUGAUUAUGAAUCGCACUCUAGCGACUACGAUGAUGAAUAUGA-CAUUGUCAACGAAGAAAAAAGUCCAAGGCUGCCCGUUGU---------------- .(((((((((((...............((((......))))....((..(((.......-)))..))..))))))....(((.....)))....)))))---------------- ( -22.90, z-score = -0.73, R) >droPer1.super_3 70259 115 + 7375914 GAUAGCUCCUCAAUUGGCGACUACGAGUCGCACUCGAGCGACUACGACGACGAUGACGAGGAACCAGUACCGGAACCGGAACUGGAACAAGAACGGAAACACAAUACGAGUCUGC (((...(((((.((((.((....((((((((......)))))).)).)).))))...))))).(((((.(((....))).))))).........(....).........)))... ( -35.90, z-score = -3.21, R) >dp4.chr2 6269916 115 + 30794189 GAUAGCUCCUCAAUUGGCGACUACGAGUCGCACUCGAGCGACUACGACGACGAUGACGAGGAACCAGUACCGGAACCGGAACUGGAACAAGAACGGAAACACAAUACGAGUCUGC (((...(((((.((((.((....((((((((......)))))).)).)).))))...))))).(((((.(((....))).))))).........(....).........)))... ( -35.90, z-score = -3.21, R) >droAna3.scaffold_13340 11248337 80 - 23697760 GACAGCUCUUCGAUUGGGGACUACGAGUCGCAUUCCAGCGACUACGAUGACGAAGA----GAACGAGGAUGAGAAUGAGAAUGA------------------------------- .....(((((((...(....)..((((((((......)))))).))....))))))----).......................------------------------------- ( -22.80, z-score = -2.25, R) >droEre2.scaffold_4770 597842 101 - 17746568 GACAGCUCUUCGAUUGGUGAUUAUGAAUCGCACUCUAGCGAUUACGACGAUGAAUAUGA-CAUUGUCAACGAAGAUAAAAACCCAAGGUUGUUAGCAGGUGU------------- ....((((((((.............((((((......))))))..(((((((.......-)))))))..))))))(((.((((...)))).)))))......------------- ( -25.90, z-score = -1.54, R) >droGri2.scaffold_14906 3875468 85 - 14172833 GACAGUUCUUCCAUUGGAGAUUAUGAGUCGCACUCAAGCGACUACGAUGAUGAGAUUGA-GGGCGUCAAUGAUGAAUGUCUGCUAU----------------------------- ((((.(((...(((((..(......((((((......)))))).((((......)))).-...)..)))))..)))))))......----------------------------- ( -21.00, z-score = -0.01, R) >consensus GACAGCUCUUCGAUUGGUGAUUAUGAAUCGCACUCUAGCGACUACGAUGAUGAAUAUGA_CAUUGUCAACGAAGAAAAAAACCCAAGGCUGCACGUUGU________________ .......................((((((((......)))))).))..................................................................... ( -7.79 = -7.61 + -0.18)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:49:25 2011