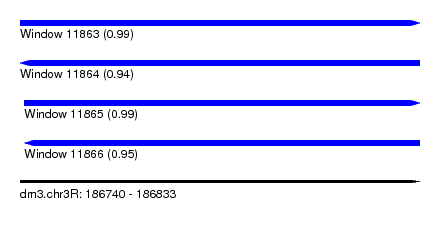
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 186,740 – 186,833 |
| Length | 93 |
| Max. P | 0.987438 |

| Location | 186,740 – 186,833 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 85.08 |
| Shannon entropy | 0.25338 |
| G+C content | 0.52560 |
| Mean single sequence MFE | -36.76 |
| Consensus MFE | -28.20 |
| Energy contribution | -28.96 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.986067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
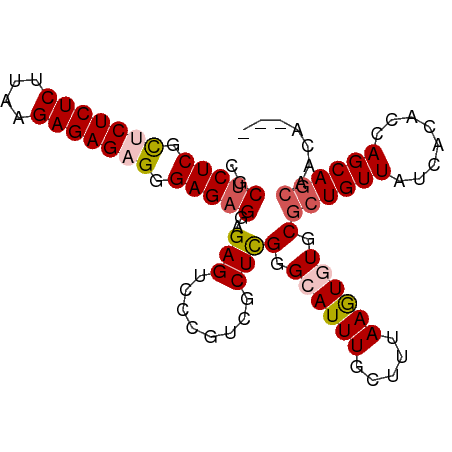
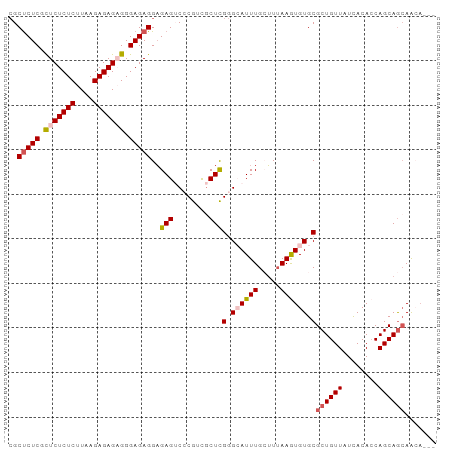
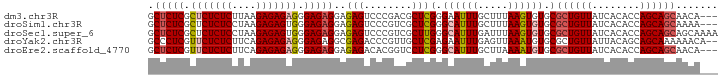
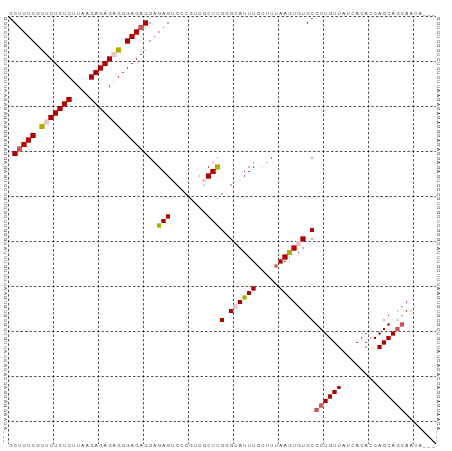
>dm3.chr3R 186740 93 + 27905053 CGCUCUCGCUCUCUCUUAAGAGAGAGGGAGAGGAGAGUCCCGACGCUCGGGAAUUUGCUUUAAGUGUGCGCUGUUAUCACACCAGCAGCAACA--- ..(((((.(((((((....))))))).)))))(((..(((((.....))))).....)))....(((..((((((........)))))).)))--- ( -38.90, z-score = -2.87, R) >droSim1.chr3R 257187 93 + 27517382 CGCUCUCGCUCUCUCCUAAGAGAGUGGGAGAGGAGAGUCCCGUCGCUCGGGCAUUUGCUUUAAGUGUGCGCUGUUAUCACACCAGCAGCAAAA--- .(((((((((((((....)))))))))))((((..(((((((.....)))).)))..))))......))((((((........))))))....--- ( -35.70, z-score = -1.98, R) >droSec1.super_6 301702 96 + 4358794 CGCUCUCGCUCUCUCCUAAGAGAGUGGGAGAGGAGAGUCCCGUCGCUUGGGCAUUUGAUUUAAGUGUGCGCUGUUAUCACACCAGCAGCAGCAAAA ..((((((((((((....)))))))))))).((......))...(((.(.(((((((...))))))).)((((((........))))))))).... ( -35.50, z-score = -1.78, R) >droYak2.chr3R 545715 93 + 28832112 -GCCCUCGUUCUCUCUUCAGAGAGAGGGAGAGGCGAGACCCGUUGCUCGAGAAUUUGAGUUAAAUGUGCGCUGUUAUUACAGCAGCAAAAAACA-- -((((((.(((((((....))))))).))).)))......((((((((((....))))))..))))((((((((....))))).))).......-- ( -35.80, z-score = -3.64, R) >droEre2.scaffold_4770 538094 93 + 17746568 CGCUCUCGUUCUCUCUUCAGAGAGAGGGAGAGGAGAGACACGGUCCUCGGGCAUUUGCUUAAAAUGUGCGCUGUUAUCACACCAGCAGCAACA--- ..(((((.(((((((....))))))).)))))(((.(((...))))))(.((((((.....)))))).)((((((........))))))....--- ( -37.90, z-score = -3.39, R) >consensus CGCUCUCGCUCUCUCUUAAGAGAGAGGGAGAGGAGAGUCCCGUCGCUCGGGCAUUUGCUUUAAGUGUGCGCUGUUAUCACACCAGCAGCAACA___ ..(((((.(((((((....))))))).)))))..(((........)))(.((((((.....)))))).)((((((........))))))....... (-28.20 = -28.96 + 0.76)
| Location | 186,740 – 186,833 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 85.08 |
| Shannon entropy | 0.25338 |
| G+C content | 0.52560 |
| Mean single sequence MFE | -32.26 |
| Consensus MFE | -24.74 |
| Energy contribution | -26.94 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.941915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
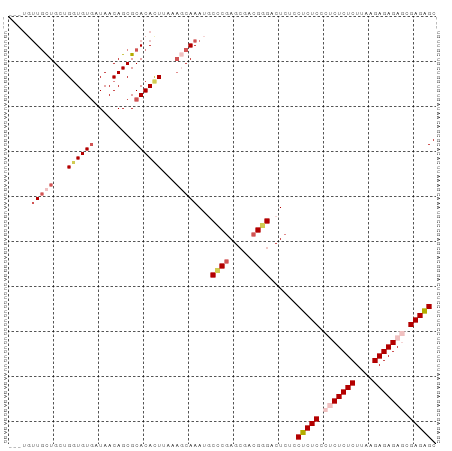
>dm3.chr3R 186740 93 - 27905053 ---UGUUGCUGCUGGUGUGAUAACAGCGCACACUUAAAGCAAAUUCCCGAGCGUCGGGACUCUCCUCUCCCUCUCUCUUAAGAGAGAGCGAGAGCG ---..(((((...((((((.........))))))...)))))..(((((.....))))).....(((((.(((((((....))))))).))))).. ( -39.90, z-score = -4.06, R) >droSim1.chr3R 257187 93 - 27517382 ---UUUUGCUGCUGGUGUGAUAACAGCGCACACUUAAAGCAAAUGCCCGAGCGACGGGACUCUCCUCUCCCACUCUCUUAGGAGAGAGCGAGAGCG ---.((((((...((((((.........))))))...))))))..((((.....)))).((((((((((((.........)).))))).))))).. ( -35.80, z-score = -2.81, R) >droSec1.super_6 301702 96 - 4358794 UUUUGCUGCUGCUGGUGUGAUAACAGCGCACACUUAAAUCAAAUGCCCAAGCGACGGGACUCUCCUCUCCCACUCUCUUAGGAGAGAGCGAGAGCG ...((.(((.((((.........))))))))).............(((.......))).((((((((((((.........)).))))).))))).. ( -28.20, z-score = -0.68, R) >droYak2.chr3R 545715 93 - 28832112 --UGUUUUUUGCUGCUGUAAUAACAGCGCACAUUUAACUCAAAUUCUCGAGCAACGGGUCUCGCCUCUCCCUCUCUCUGAAGAGAGAACGAGGGC- --.(((...(((.(((((....)))))))).....))).......((((.....))))....(((.(((..((((((....))))))..))))))- ( -27.70, z-score = -1.88, R) >droEre2.scaffold_4770 538094 93 - 17746568 ---UGUUGCUGCUGGUGUGAUAACAGCGCACAUUUUAAGCAAAUGCCCGAGGACCGUGUCUCUCCUCUCCCUCUCUCUGAAGAGAGAACGAGAGCG ---..(((((...((((((.........))))))...)))))......((((((...))).)))(((((..((((((....))))))..))))).. ( -29.70, z-score = -1.78, R) >consensus ___UGUUGCUGCUGGUGUGAUAACAGCGCACACUUAAAGCAAAUGCCCGAGCGACGGGACUCUCCUCUCCCUCUCUCUUAAGAGAGAGCGAGAGCG .....(((((...((((((.........))))))...)))))...((((.....))))......(((((.(((((((....))))))).))))).. (-24.74 = -26.94 + 2.20)

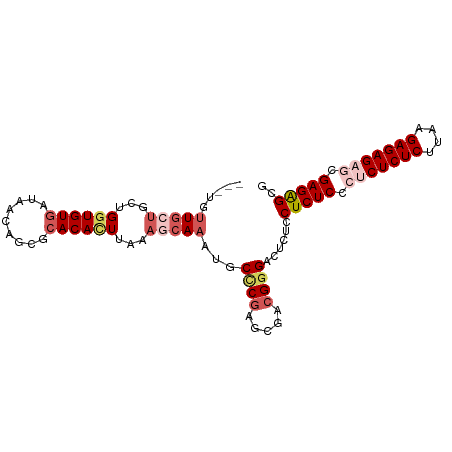
| Location | 186,741 – 186,833 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 85.35 |
| Shannon entropy | 0.24844 |
| G+C content | 0.52160 |
| Mean single sequence MFE | -36.70 |
| Consensus MFE | -28.20 |
| Energy contribution | -28.96 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.987438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
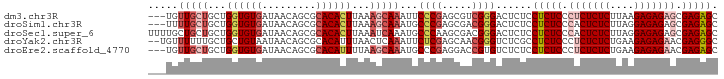
>dm3.chr3R 186741 92 + 27905053 GCUCUCGCUCUCUCUUAAGAGAGAGGGAGAGGAGAGUCCCGACGCUCGGGAAUUUGCUUUAAGUGUGCGCUGUUAUCACACCAGCAGCAACA--- .(((((.(((((((....))))))).)))))(((..(((((.....))))).....)))....(((..((((((........)))))).)))--- ( -38.90, z-score = -2.98, R) >droSim1.chr3R 257188 92 + 27517382 GCUCUCGCUCUCUCCUAAGAGAGUGGGAGAGGAGAGUCCCGUCGCUCGGGCAUUUGCUUUAAGUGUGCGCUGUUAUCACACCAGCAGCAAAA--- (((((((((((((....)))))))))))((((..(((((((.....)))).)))..))))......))((((((........))))))....--- ( -35.40, z-score = -1.98, R) >droSec1.super_6 301703 95 + 4358794 GCUCUCGCUCUCUCCUAAGAGAGUGGGAGAGGAGAGUCCCGUCGCUUGGGCAUUUGAUUUAAGUGUGCGCUGUUAUCACACCAGCAGCAGCAAAA .((((((((((((....)))))))))))).((......))...(((.(.(((((((...))))))).)((((((........))))))))).... ( -35.50, z-score = -1.86, R) >droYak2.chr3R 545715 93 + 28832112 GCCCUCGUUCUCUCUUCAGAGAGAGGGAGAGGCGAGACCCGUUGCUCGAGAAUUUGAGUUAAAUGUGCGCUGUUAUUACAGCAGCAAAAAACA-- ((((((.(((((((....))))))).))).)))......((((((((((....))))))..))))((((((((....))))).))).......-- ( -35.80, z-score = -3.64, R) >droEre2.scaffold_4770 538095 92 + 17746568 GCUCUCGUUCUCUCUUCAGAGAGAGGGAGAGGAGAGACACGGUCCUCGGGCAUUUGCUUAAAAUGUGCGCUGUUAUCACACCAGCAGCAACA--- .(((((.(((((((....))))))).)))))(((.(((...))))))(.((((((.....)))))).)((((((........))))))....--- ( -37.90, z-score = -3.47, R) >consensus GCUCUCGCUCUCUCUUAAGAGAGAGGGAGAGGAGAGUCCCGUCGCUCGGGCAUUUGCUUUAAGUGUGCGCUGUUAUCACACCAGCAGCAACA___ .(((((.(((((((....))))))).)))))..(((........)))(.((((((.....)))))).)((((((........))))))....... (-28.20 = -28.96 + 0.76)
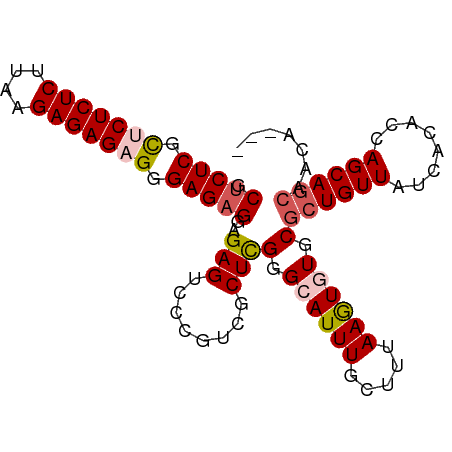
| Location | 186,741 – 186,833 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 85.35 |
| Shannon entropy | 0.24844 |
| G+C content | 0.52160 |
| Mean single sequence MFE | -32.26 |
| Consensus MFE | -24.74 |
| Energy contribution | -26.94 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.950078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
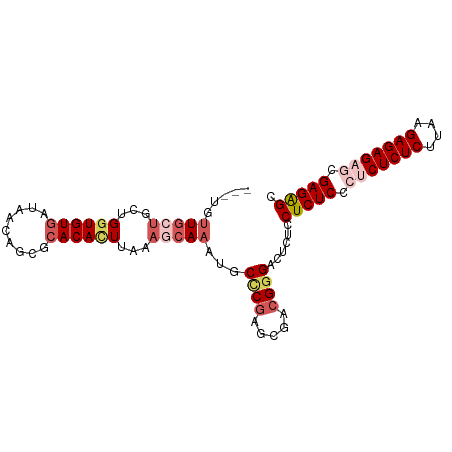
>dm3.chr3R 186741 92 - 27905053 ---UGUUGCUGCUGGUGUGAUAACAGCGCACACUUAAAGCAAAUUCCCGAGCGUCGGGACUCUCCUCUCCCUCUCUCUUAAGAGAGAGCGAGAGC ---..(((((...((((((.........))))))...)))))..(((((.....))))).....(((((.(((((((....))))))).))))). ( -39.90, z-score = -4.16, R) >droSim1.chr3R 257188 92 - 27517382 ---UUUUGCUGCUGGUGUGAUAACAGCGCACACUUAAAGCAAAUGCCCGAGCGACGGGACUCUCCUCUCCCACUCUCUUAGGAGAGAGCGAGAGC ---.((((((...((((((.........))))))...))))))..((((.....)))).((((((((((((.........)).))))).))))). ( -35.80, z-score = -2.91, R) >droSec1.super_6 301703 95 - 4358794 UUUUGCUGCUGCUGGUGUGAUAACAGCGCACACUUAAAUCAAAUGCCCAAGCGACGGGACUCUCCUCUCCCACUCUCUUAGGAGAGAGCGAGAGC ...((.(((.((((.........))))))))).............(((.......))).((((((((((((.........)).))))).))))). ( -28.20, z-score = -0.74, R) >droYak2.chr3R 545715 93 - 28832112 --UGUUUUUUGCUGCUGUAAUAACAGCGCACAUUUAACUCAAAUUCUCGAGCAACGGGUCUCGCCUCUCCCUCUCUCUGAAGAGAGAACGAGGGC --.(((...(((.(((((....)))))))).....))).......((((.....))))....(((.(((..((((((....))))))..)))))) ( -27.70, z-score = -1.88, R) >droEre2.scaffold_4770 538095 92 - 17746568 ---UGUUGCUGCUGGUGUGAUAACAGCGCACAUUUUAAGCAAAUGCCCGAGGACCGUGUCUCUCCUCUCCCUCUCUCUGAAGAGAGAACGAGAGC ---..(((((...((((((.........))))))...)))))......((((((...))).)))(((((..((((((....))))))..))))). ( -29.70, z-score = -1.87, R) >consensus ___UGUUGCUGCUGGUGUGAUAACAGCGCACACUUAAAGCAAAUGCCCGAGCGACGGGACUCUCCUCUCCCUCUCUCUUAAGAGAGAGCGAGAGC .....(((((...((((((.........))))))...)))))...((((.....))))......(((((.(((((((....))))))).))))). (-24.74 = -26.94 + 2.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:49:24 2011