
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 143,749 – 143,867 |
| Length | 118 |
| Max. P | 0.914415 |

| Location | 143,749 – 143,867 |
|---|---|
| Length | 118 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.32 |
| Shannon entropy | 0.55324 |
| G+C content | 0.59185 |
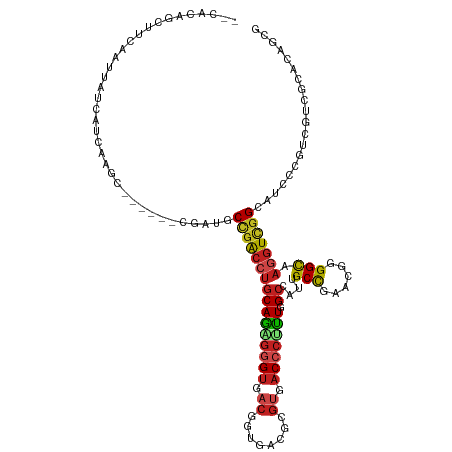
| Mean single sequence MFE | -41.44 |
| Consensus MFE | -22.86 |
| Energy contribution | -23.01 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.914415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
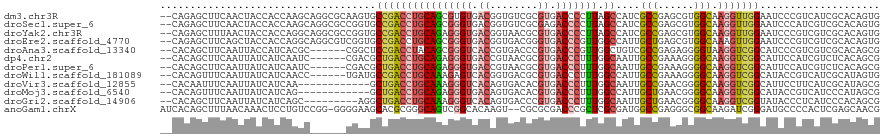
>dm3.chr3R 143749 118 - 27905053 --CAGAGCUUCAACUACCACCAAGCAGGCGCAAGUGCCGACCUGCAGCGUGUGACGGUGUCGCGUGACCCCUUAGCCAUCGCCGAGCGUGGCAAGGUUGGAAUCCCGUCAUCGCACAGUG --...............(((...((((((((....).)).))))).(((.(((((((.(((....))).((((.((((.(((...)))))))))))........))))))))))...))) ( -40.80, z-score = -0.29, R) >droSec1.super_6 262380 118 - 4358794 --CAGAGCUUCAACUACCACCAAGCAGGCGCCGGUGCCGACCUGCAGCGGGUGACGGUGUCGCGAGACCCCUUAGCCAUCGCCGAGCGUGGCAAGGUUGGAAUCCCAUCGUCGCACAGUG --....((((...........))))..(((.((((((((((((((.((.(((((.((((((....)))......))).)))))..))...)).))))))).....))))).)))...... ( -42.80, z-score = -0.04, R) >droYak2.chr3R 503847 118 - 28832112 --CAGAGCUUUAACUACCACCAGGCAGGCGCCGGUGCCGACCUGCAGAGGGUGACGGUAACGCGUGACCCCUUAGCCAUCGCCGAGCGUGGCAAGGUUGGAAUCCCGUCGUCGCACAGUG --...............(((...((.((((.(((..(((((((..((.((((.(((......))).))))))..((((.(((...))))))).)))))))....))).))))))...))) ( -45.50, z-score = -0.67, R) >droEre2.scaffold_4770 494623 118 - 17746568 --CAGAGCUUCAGCUACCACCAGGCAGGCGUCGGUGCCGACCUGCAGCGGGUGACGGUGACGGGUGACCCGUUGGCCAUUGCUGAGCGUGGCAAAGUUGGAAUCCCGUCGUCGCACAGUG --...(((....)))..(((...((.((((.(((..(((((...((((((((.(((....)..)).))))))))(((((.((...)))))))...)))))....))).))))))...))) ( -48.10, z-score = -0.16, R) >droAna3.scaffold_13340 11149143 112 - 23697760 --CACAGCUUCAAUUACCAUCACGC------CGGCUCCGACCUACAGCGGGUCACCGUGACCCGUGACCCGUUGGCUGUCGCCGAGAGGGGUAAGGUCGGCAUCCCGUCGUCGCACAGCG --...................(((.------(((..(((((((((.(((((((.....)))))))..(((.(((((....)))))..))))).)))))))....))).)))(((...))) ( -44.30, z-score = -1.31, R) >dp4.chr2 6009268 112 + 30794189 --CACAGCUUCAAUUAUCAUCAAUC------CGACGCUGACCUGCAGAGGGUGACCGUAACGCGUGACCCUUUGGCAAUUGCCGAAAGGGGCAAGGUCGGCAUUCCAUCGUCUCACAGCG --....(((..........((....------.)).((((((((((...((....))...........(((((((((....))).)))))))).))))))))...............))). ( -37.50, z-score = -1.68, R) >droPer1.super_6 6055492 112 + 6141320 --CACAGCUUCAAUUAUCAUCAAUC------CGACGCUGACCUGCAGAGGGUGACCGUAACGCGUGACCCUUUGGCAAUUGCCGAAAGGGGCAAGGUCGGCAUUCCAUCGUCUCACAGCG --....(((..........((....------.)).((((((((((...((....))...........(((((((((....))).)))))))).))))))))...............))). ( -37.50, z-score = -1.68, R) >droWil1.scaffold_181089 2586371 112 + 12369635 --CACAGUUUCAAUUAUCAUCAACC------UGAUGCCGACCUGCAAAGAGUCACGGUGACGCGUGACCCUUUGGCCAUUGCCGAAAGGGGCAAGGUCGGCAUACCGUCAUCGCAUAGUG --(((....................------..((((((((((.(((((.((((((......)))))).)))))(((....((....))))).))))))))))..((....))....))) ( -41.60, z-score = -2.99, R) >droVir3.scaffold_12855 2843466 106 - 10161210 --CACAAUUUCAAUUAUCAUCAA------------GCUGACCUGCAAAGGGUCACAGUGACACGUGACCCUUUGGCAAUUGCCGAACGGGGCAAGGUCGGCAUUCCUUCAUCGCAUAGCG --.....................------------((((((((((((((((((((........)))))))))).))).(((((......))))))))))))..........(((...))) ( -39.70, z-score = -4.35, R) >droMoj3.scaffold_6540 23778474 106 + 34148556 --CACAGUUUCAAUUAUCAUCAG------------GCUGACCUGCAGAGGGUGACAGUGACACGUGACCCUUUGGCCAUUGCUGAACGGGGCAAGGUCGGCAUACCGUCAUCCCAUAGCG --....(((..............------------((((((((.((((((((.((........)).))))))))(((.(((.....)))))).))))))))...............))). ( -33.75, z-score = -0.72, R) >droGri2.scaffold_14906 10348132 109 + 14172833 --CACAGCUUCAAUUAUCAUCAGC---------AGGCUGACCUGCAAAGGGUCACAGUGACCCGUGACCCUUUGGCAAUUGCUGAACGGGGCAAGGUCGGUAUACCCUCAUCCCACAGCG --....(((.............((---------(((....)))))((((((((((........)))))))))))))....((((...((((..(((.........)))..)))).)))). ( -39.30, z-score = -2.48, R) >anoGam1.chrX 7649152 117 - 22145176 AUCACAGCUUUAACAAACUCCUGUCCGG-GGGGAAGCACGCGGGCAGUCGGCACAAGU--CGCGCGACCCGCUCGCGAUGGCCGAGGGCGGCAAGAUCGGGAUGCCCCACUCGAGCAACG ......(((((......(((((....))-)))))))).(((((((.((((((......--.)).))))..)))))))...((((((((.((((.........)))))).)))).)).... ( -46.40, z-score = -0.43, R) >consensus __CACAGCUUCAAUUAUCAUCAAGC______CGAUGCCGACCUGCAGAGGGUGACGGUGACGCGUGACCCUUUGGCCAUUGCCGAACGGGGCAAGGUCGGCAUCCCGUCGUCGCACAGCG ....................................((((((((((((((((.((........)).))))))).))....(((......))).))))))).................... (-22.86 = -23.01 + 0.15)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:49:19 2011