
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 137,230 – 137,322 |
| Length | 92 |
| Max. P | 0.598042 |

| Location | 137,230 – 137,322 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 57.00 |
| Shannon entropy | 0.75860 |
| G+C content | 0.45683 |
| Mean single sequence MFE | -23.51 |
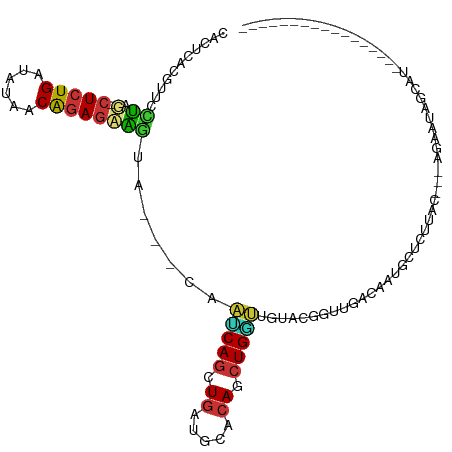
| Consensus MFE | -7.33 |
| Energy contribution | -7.12 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.598042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
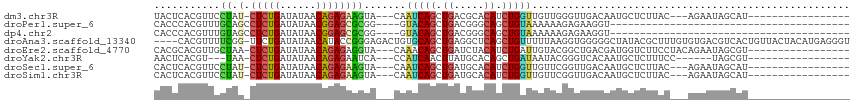
>dm3.chr3R 137230 92 - 27905053 UACUCACGUUCCUAU-CUCUGAUAUAACAGAGAAGUA---CAAUCAGCUGACGCACAUCUGGUUGUUGGGUUGACAAUGCUCUUAC---AGAAUAGCAU----------------- .......((((((((-(((((......))))))...(---(((((((.((.....)).))))))))))))..))).(((((.((..---..)).)))))----------------- ( -23.00, z-score = -1.32, R) >droPer1.super_6 6049326 72 + 6141320 CACCCACGUUUGCAGCCUCUGAUAUAACGGAGCGCGG----GUACAGCUGACGGGCAGCUGUAAAAAAGAGAAGGU---------------------------------------- .((((..(....).(((((((......))))).))))----))(((((((.....)))))))..............---------------------------------------- ( -23.90, z-score = -1.83, R) >dp4.chr2 6003077 72 + 30794189 CACCCACGUUUGUAGCCUCUGAUAUAACGGAGCGCGG----GUACAGCUGACGGGCAGCUGUAAAAAAGAGAAGGU---------------------------------------- .((((((....)).(((((((......))))).))))----))(((((((.....)))))))..............---------------------------------------- ( -23.50, z-score = -1.91, R) >droAna3.scaffold_13340 11143025 111 - 23697760 ----CACGUUUUCGG-UUCUGAUAUAACAUACCGGGAGACUGUGCAGCUGAGGCUCAGCUGUUUUUAAGGUGGGGGCUAUACGCUUUGUGUGACGUCACUGUUACUACAUGAGGGU ----.........((-((((.((.........(((....))).(((((((.....))))))).......)).))))))..((.((((((((((((....)))))).))).))).)) ( -31.00, z-score = -0.09, R) >droEre2.scaffold_4770 487864 95 - 17746568 CACGCACGUUGCUAA-CUCUGAUAUAACAGAGAGGUA---CAAACAGCUGAUCUACAUCUGAUUGUACGGCUGACGAUGGUCUUCCUACAGAAUAGCGU----------------- .((((.(((((((..-(((((......)))))..(((---(((.(((.((.....)).))).))))))))).))))..((....)).........))))----------------- ( -28.00, z-score = -2.34, R) >droYak2.chr3R 496537 86 - 28832112 AACUCACGU---UAA-CUCUGAUAUAACAGAGAAUCA---CCAUCAACUUAUGCACAGCUGAUAAUACGGGUCACAAUGCUCUUUCC------UAGCGU----------------- .....((((---((.-(((((......))))).....---............(((..(.((((.......)))))..))).......------))))))----------------- ( -14.50, z-score = -0.85, R) >droSec1.super_6 256076 92 - 4358794 CACUCACGUUCCUAU-CUCUGAUAUAACAGAGAAGUA---CAAUCAGCUGAUGCACAUCUGGUUGUUCGGUUGACAAUGCUCUUAC---AGAAUAGCAU----------------- .......(((((..(-(((((......)))))).(.(---(((((((.((.....)).)))))))).)))..))).(((((.((..---..)).)))))----------------- ( -22.10, z-score = -1.09, R) >droSim1.chr3R 209625 92 - 27517382 CACUCACGUUCCUAU-CUCUGAUAUAACAGAGAAGUA---CAAUCAGCUGAUGCACAUCUGGUUGUUCGGUUGACAAUGCUCUUAC---AGAAUAGCAU----------------- .......(((((..(-(((((......)))))).(.(---(((((((.((.....)).)))))))).)))..))).(((((.((..---..)).)))))----------------- ( -22.10, z-score = -1.09, R) >consensus CACUCACGUUCCUAG_CUCUGAUAUAACAGAGAAGUA___CAAUCAGCUGAUGCACAGCUGGUUGUACGGUUGACAAUGCUCUUAC___AGAAUAGCAU_________________ ................(((((......)))))..........(((((.((.....)).)))))..................................................... ( -7.33 = -7.12 + -0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:49:18 2011