| Sequence ID | dm3.chr3R |
|---|---|
| Location | 46,630 – 46,745 |
| Length | 115 |
| Max. P | 0.984223 |

| Location | 46,630 – 46,745 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.95 |
| Shannon entropy | 0.44372 |
| G+C content | 0.59963 |
| Mean single sequence MFE | -38.60 |
| Consensus MFE | -25.82 |
| Energy contribution | -27.78 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.984223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
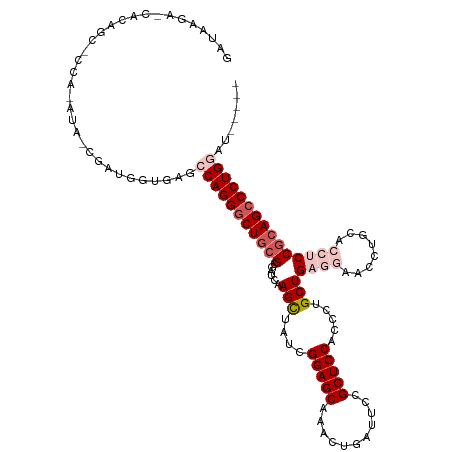
>dm3.chr3R 46630 115 + 27905053 GAUAAGAGCACAGCGCCACAUAUCGAUGGUGAGCCAGGCCUGCGCGAUCAAGCUAUCGGAGCAAACUGAUUCCGCUCCACCCUGCUGAGGAACCUGCUCCUCCGCAGCCCUGGAU----- .............(((((........)))))..(((((.(((((......(((....(((((...........))))).....)))(((((......)))))))))).)))))..----- ( -38.90, z-score = -1.07, R) >droEre2.scaffold_4770 404958 97 + 17746568 -----------------------CGAUGGCGCGCCAGGGCUGCGGCAUCAAGUUAUCGGAGCCAACUGAUUCCGCUCCCACCUGCUGCCGAGCCUGCACCUCCGCAGCCCUGCACAUUCU -----------------------.......(.((.((((((((((((.((.((....(((((...........))))).)).)).))))(((.......))).)))))))))).)..... ( -36.00, z-score = -1.05, R) >droYak2.chr3R 409981 97 + 28832112 -----------------------CGAUUGUUCGCCAGGGCUACGGGAUCAAGUUAUCGGAGCCAACAGCUUCCGCUCCCACCUGCUGCCGAACCUGGACCUCCGCAGCCCUGCACAUUCA -----------------------.((.(((..((.((((((.((((.((.((...(((((((....(((....))).......))).))))..)).)).).))).))))))))))).)). ( -32.20, z-score = -1.41, R) >droSec1.super_6 174439 114 + 4358794 GAUAAGAUCACAGCACCAAAUAUCGAUGUUGAGCCAGGGCUGGGCGAUCAAGCUAUCGGAGCAAACUGAUUCCGCUCCACCCCGCUGAG-AACCAGCACCUCCGCAGCCCUGGAU----- ..........(((((.(.......).)))))..(((((((((((.(...........(((((...........))))).....((((..-...))))..).)).)))))))))..----- ( -39.60, z-score = -2.59, R) >droSim1.chr3R 129208 114 + 27517382 GAUAAGAUCACAGCACCAAAUACCGAUGUUGAGCCAGGGCUGCGCGAUCAAGCUAUCGGAGCAAACUGGUUCCGCUCCGCCCCGCUGAG-AACCAGCACCUCCGCAGCCCUGGAU----- ..........(((((.(.......).)))))..((((((((((((......))....((((....((((((((((........)).).)-))))))...))))))))))))))..----- ( -46.30, z-score = -4.28, R) >consensus GAUAAGA_CACAGC_CCA_AUA_CGAUGGUGAGCCAGGGCUGCGCGAUCAAGCUAUCGGAGCAAACUGAUUCCGCUCCACCCUGCUGAGGAACCUGCACCUCCGCAGCCCUGGAU_____ .................................(((((((((((......(((....(((((...........))))).....)))((((........)))))))))))))))....... (-25.82 = -27.78 + 1.96)

| Location | 46,630 – 46,745 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.95 |
| Shannon entropy | 0.44372 |
| G+C content | 0.59963 |
| Mean single sequence MFE | -47.48 |
| Consensus MFE | -30.80 |
| Energy contribution | -33.12 |
| Covariance contribution | 2.32 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.980530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
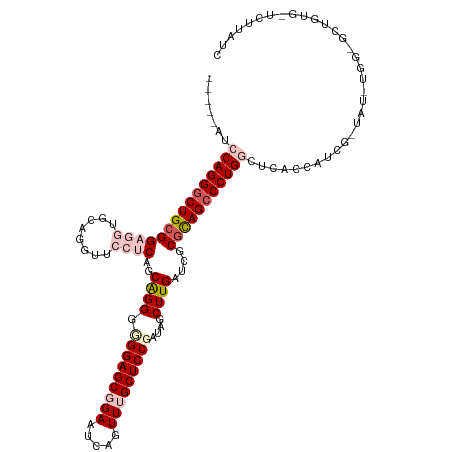
>dm3.chr3R 46630 115 - 27905053 -----AUCCAGGGCUGCGGAGGAGCAGGUUCCUCAGCAGGGUGGAGCGGAAUCAGUUUGCUCCGAUAGCUUGAUCGCGCAGGCCUGGCUCACCAUCGAUAUGUGGCGCUGUGCUCUUAUC -----..(((((.(((((((((((....))))))..((((.(((((((((.....)))))))))....))))....))))).))))).........((((...(((.....)))..)))) ( -45.50, z-score = -0.44, R) >droEre2.scaffold_4770 404958 97 - 17746568 AGAAUGUGCAGGGCUGCGGAGGUGCAGGCUCGGCAGCAGGUGGGAGCGGAAUCAGUUGGCUCCGAUAACUUGAUGCCGCAGCCCUGGCGCGCCAUCG----------------------- .....((((((((((((.(((.......)))((((.(((((.(((((.((.....)).)))))....))))).)))))))))))).)))).......----------------------- ( -48.10, z-score = -2.27, R) >droYak2.chr3R 409981 97 - 28832112 UGAAUGUGCAGGGCUGCGGAGGUCCAGGUUCGGCAGCAGGUGGGAGCGGAAGCUGUUGGCUCCGAUAACUUGAUCCCGUAGCCCUGGCGAACAAUCG----------------------- .((.(((.(((((((((((.((((.((..((((.(((....((.(((....))).)).)))))))...)).)))))))))))))))....))).)).----------------------- ( -49.20, z-score = -4.19, R) >droSec1.super_6 174439 114 - 4358794 -----AUCCAGGGCUGCGGAGGUGCUGGUU-CUCAGCGGGGUGGAGCGGAAUCAGUUUGCUCCGAUAGCUUGAUCGCCCAGCCCUGGCUCAACAUCGAUAUUUGGUGCUGUGAUCUUAUC -----..((((((((((.((((.......)-))).)).((((((((((((.....))))))))(((......)))))))))))))))(.((.((((((...)))))).)).)........ ( -44.20, z-score = -1.14, R) >droSim1.chr3R 129208 114 - 27517382 -----AUCCAGGGCUGCGGAGGUGCUGGUU-CUCAGCGGGGCGGAGCGGAACCAGUUUGCUCCGAUAGCUUGAUCGCGCAGCCCUGGCUCAACAUCGGUAUUUGGUGCUGUGAUCUUAUC -----..((((((((((((((..(((((((-((..((........)))))))))))...))))....((......)))))))))))).....((.((((((...))))))))........ ( -50.40, z-score = -2.33, R) >consensus _____AUCCAGGGCUGCGGAGGUGCAGGUUCCUCAGCAGGGGGGAGCGGAAUCAGUUUGCUCCGAUAGCUUGAUCGCGCAGCCCUGGCUCACCAUCG_UAU_UGG_GCUGUG_UCUUAUC .......(((((((((((((((........))))..((((.(((((((((.....)))))))))....))))....)))))))))))................................. (-30.80 = -33.12 + 2.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:49:15 2011