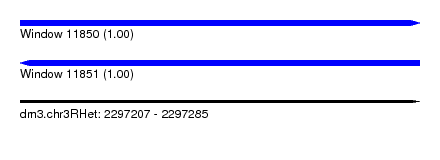
| Sequence ID | dm3.chr3RHet |
|---|---|
| Location | 2,297,207 – 2,297,285 |
| Length | 78 |
| Max. P | 0.998868 |

| Location | 2,297,207 – 2,297,285 |
|---|---|
| Length | 78 |
| Sequences | 4 |
| Columns | 78 |
| Reading direction | forward |
| Mean pairwise identity | 70.37 |
| Shannon entropy | 0.48438 |
| G+C content | 0.37495 |
| Mean single sequence MFE | -17.62 |
| Consensus MFE | -14.05 |
| Energy contribution | -13.93 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.12 |
| SVM RNA-class probability | 0.997540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
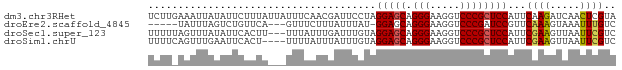
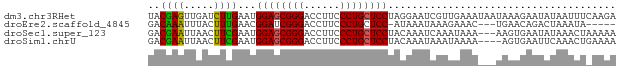
>dm3.chr3RHet 2297207 78 + 2517507 UCUUGAAAUUAUAUUCUUUAUUAUUUCAACGAUUCCUAGGAGCAGGGAAGGUCCCGCUCCAUUCAAGAUCAACUCGUA (((((((((.(((.....))).))))))).))......(((((.(((.....)))))))).................. ( -20.00, z-score = -2.62, R) >droEre2.scaffold_4845 22507504 69 - 22589142 -----UAUUUAGUCUGUUCA---GUUUCUUUAUUUAU-GGAGCAGGGAAGGUCCCGAUCCGUUCAAAGUAAAUUUGUC -----..............(---((((((((....((-(((...(((.....)))..)))))..)))).))))).... ( -13.30, z-score = -1.28, R) >droSec1.super_123 70417 75 + 76498 UUUUUAGUUUAUAUUCACUU---UUUAUUUGAUUUGUAGGAGCAGGGAAGGUCCCGCUCCAUUCGAAGUUAAUUCGUC ....................---...............(((((.(((.....))))))))...((((.....)))).. ( -16.70, z-score = -2.06, R) >droSim1.chrU 10701465 74 - 15797150 UUUUCAGUUUGAAUUCACU----UUUUAUUUAUUUGUAGGAGCAGGGAAGGUCCCGCUCCAUUCGAAGUUAAUUCGUC ..........(((((.(((----((.(((......)))(((((.(((.....))))))))....))))).)))))... ( -20.50, z-score = -3.54, R) >consensus UUUU_AAUUUAUAUUCACUA___UUUUAUUGAUUUGUAGGAGCAGGGAAGGUCCCGCUCCAUUCAAAGUUAAUUCGUC ......................................(((((.(((.....))))))))...((((.....)))).. (-14.05 = -13.93 + -0.13)
| Location | 2,297,207 – 2,297,285 |
|---|---|
| Length | 78 |
| Sequences | 4 |
| Columns | 78 |
| Reading direction | reverse |
| Mean pairwise identity | 70.37 |
| Shannon entropy | 0.48438 |
| G+C content | 0.37495 |
| Mean single sequence MFE | -17.33 |
| Consensus MFE | -15.89 |
| Energy contribution | -15.32 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.53 |
| SVM RNA-class probability | 0.998868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
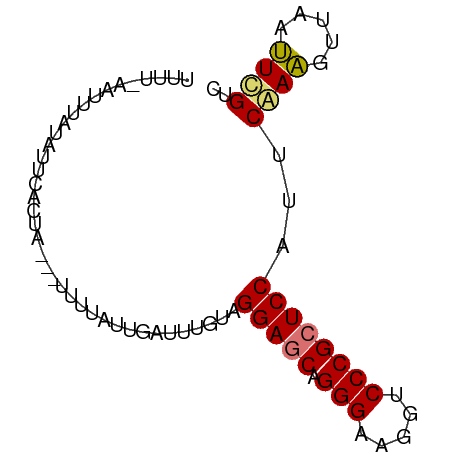
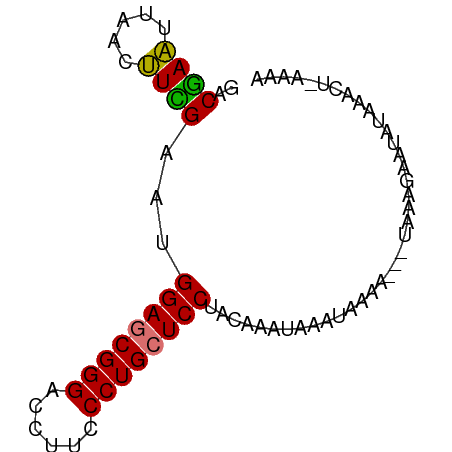
>dm3.chr3RHet 2297207 78 - 2517507 UACGAGUUGAUCUUGAAUGGAGCGGGACCUUCCCUGCUCCUAGGAAUCGUUGAAAUAAUAAAGAAUAUAAUUUCAAGA .((((.....(((((...((((((((......))))))))))))).))))((((((.(((.....))).))))))... ( -22.30, z-score = -2.86, R) >droEre2.scaffold_4845 22507504 69 + 22589142 GACAAAUUUACUUUGAACGGAUCGGGACCUUCCCUGCUCC-AUAAAUAAAGAAAC---UGAACAGACUAAAUA----- .....(((((.((((...(((.((((......)))).)))-........((...)---)...)))).))))).----- ( -10.40, z-score = -1.57, R) >droSec1.super_123 70417 75 - 76498 GACGAAUUAACUUCGAAUGGAGCGGGACCUUCCCUGCUCCUACAAAUCAAAUAAA---AAGUGAAUAUAAACUAAAAA ..((((.....))))...((((((((......))))))))...............---.(((........)))..... ( -17.90, z-score = -4.24, R) >droSim1.chrU 10701465 74 + 15797150 GACGAAUUAACUUCGAAUGGAGCGGGACCUUCCCUGCUCCUACAAAUAAAUAAAA----AGUGAAUUCAAACUGAAAA ...(((((.((((.....((((((((......))))))))..............)----))).))))).......... ( -18.71, z-score = -4.25, R) >consensus GACGAAUUAACUUCGAAUGGAGCGGGACCUUCCCUGCUCCUACAAAUAAAUAAAA___UAAAGAAUAUAAACU_AAAA ..((((.....))))...((((((((......))))))))...................................... (-15.89 = -15.32 + -0.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:49:11 2011