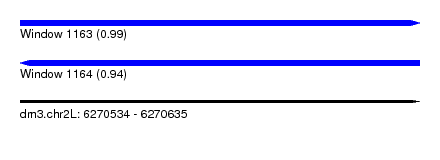
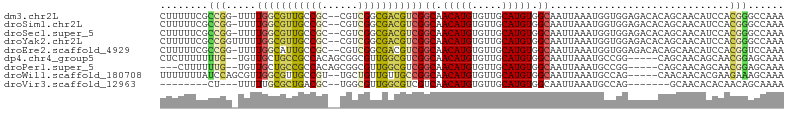
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,270,534 – 6,270,635 |
| Length | 101 |
| Max. P | 0.989668 |

| Location | 6,270,534 – 6,270,635 |
|---|---|
| Length | 101 |
| Sequences | 9 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 76.90 |
| Shannon entropy | 0.45999 |
| G+C content | 0.52971 |
| Mean single sequence MFE | -36.57 |
| Consensus MFE | -20.98 |
| Energy contribution | -19.75 |
| Covariance contribution | -1.23 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.989668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
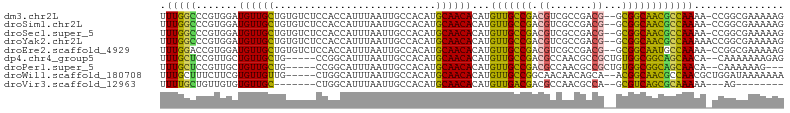
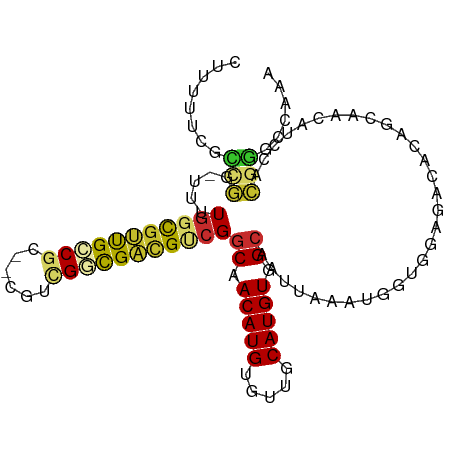
>dm3.chr2L 6270534 101 + 23011544 UUUGGCCCGUGGAUGUUGCUGUGUCUCCACCAUUUAAUUGCCACAUGCAACACAUGUUGCCGACGUCGCCGACG--GCGGCAACGCCAAAA-CCGGCGAAAAAG .(((((.((((..((((((((((.(..............).)))).))))))))))..)))))..((((((..(--(((....))))....-.))))))..... ( -38.24, z-score = -2.41, R) >droSim1.chr2L 6068421 101 + 22036055 UUUGGCCCGUGGAUGUUGCUGUGUCUCCACCAUUUAAUUGCCACAUGCAACACAUGUUGCCGACGUCGCCGACG--GCGGCAACGCCAAAA-CCGGCGAAAAAG .(((((.((((..((((((((((.(..............).)))).))))))))))..)))))..((((((..(--(((....))))....-.))))))..... ( -38.24, z-score = -2.41, R) >droSec1.super_5 4341528 101 + 5866729 UUUGGCCCGUGGAUGUUGCUGUGUCUCCACCAUUUAAUUGCCACAUGCAACACAUGUUGCCGACGUCGCCGACG--GCGGCAACGCCAAAA-CCGGCGAAAAAG .(((((.((((..((((((((((.(..............).)))).))))))))))..)))))..((((((..(--(((....))))....-.))))))..... ( -38.24, z-score = -2.41, R) >droYak2.chr2L 15686787 102 - 22324452 UUUGGCCCGUGGAUGUUGCUGUGUCUCCACCAUUUAAUUGCCACAUGCAACACAUGUUGCCGACGUCGCCGACG--GCGGCAACGCCAAAAACCGGCGAAAAAG .(((((.((((..((((((((((.(..............).)))).))))))))))..)))))..((((((..(--(((....))))......))))))..... ( -37.64, z-score = -2.26, R) >droEre2.scaffold_4929 15197507 101 + 26641161 UUUGGACCGUGGAUGUUGCUGUGUCUCCACCAUUUAAUUGCCACAUGCAACACAUGUUGCCGACGUCGCCGACG--GCGGCAAUGCCAAAA-CCGGCGAAAAAG (((.(.((((((.((((((((((.(..............).)))).))))))...(((((((.((((...))))--.))))))).)))...-.)))).)))... ( -31.64, z-score = -0.72, R) >dp4.chr4_group5 192735 97 - 2436548 UUUGCUCCGUUGCUGUUGCUG-----CCGGCAUUUAAUUGCCACAUGCAACACAUGUUGCCGACGCCAACGCCGCUGUGGCGGCAGCAACA--CAAAAAAAGAG ....(((..(((.((((((((-----((((((......))))....(((((....)))))....((((.((....))))))))))))))))--))).....))) ( -38.70, z-score = -2.69, R) >droPer1.super_5 191705 94 - 6813705 UUUGCUCCGUUGCUGUUGCUG-----CCGGCAUUUAAUUGCCACAUGCAACACAUGUUGCCGACGCCAACGCCGCUGUGGCGGCAGCAACA--CAAAAAAG--- .........(((.((((((((-----((((((......))))....(((((....)))))....((((.((....))))))))))))))))--))).....--- ( -37.80, z-score = -2.61, R) >droWil1.scaffold_180708 5202437 97 - 12563649 UUUGCUUUCUUCGUGUUGUUG-----CUGGCAUUUAAUUGCCACAUGCAACACAUGUUGCCGGCAACAACAGCA--ACGGCAACGCCAACGCUGGAUAAAAAAA (((...(((..((((((((((-----((((((......))))....(((((....))))).)))))))))....--..(((...))).)))..)))...))).. ( -31.80, z-score = -2.03, R) >droVir3.scaffold_12963 2918981 84 + 20206255 UUUUGCUGUUGUGUGUUGC-------CUGGCAUUUAAUUGCCACAUGCAACACAUGUUGACGACGCCAACGCCA--GCGUCAGCGCAAAAA---AG-------- ((((((....(((((((((-------.(((((......)))))...)))))))))(((((((..((....))..--.))))))))))))).---..-------- ( -36.80, z-score = -4.46, R) >consensus UUUGGCCCGUGGAUGUUGCUGUGUCUCCACCAUUUAAUUGCCACAUGCAACACAUGUUGCCGACGUCGCCGACG__GCGGCAACGCCAAAA_CCGGCGAAAAAG .(((((.......((((((...........................))))))...(((((((.((.......))...))))))))))))............... (-20.98 = -19.75 + -1.23)
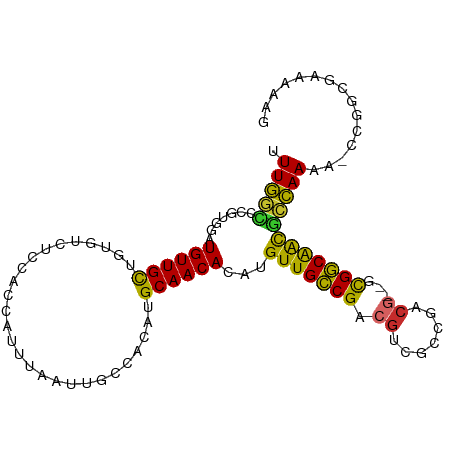
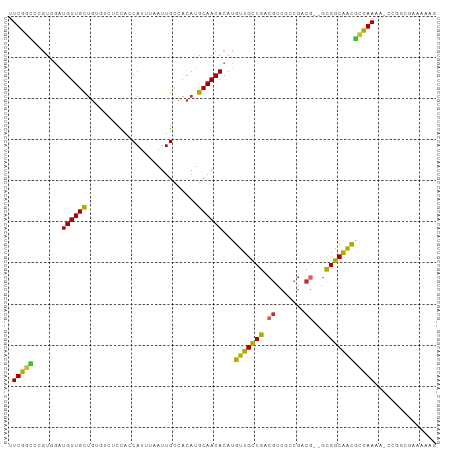
| Location | 6,270,534 – 6,270,635 |
|---|---|
| Length | 101 |
| Sequences | 9 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 76.90 |
| Shannon entropy | 0.45999 |
| G+C content | 0.52971 |
| Mean single sequence MFE | -39.12 |
| Consensus MFE | -21.46 |
| Energy contribution | -21.24 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.57 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.941641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6270534 101 - 23011544 CUUUUUCGCCGG-UUUUGGCGUUGCCGC--CGUCGGCGACGUCGGCAACAUGUGUUGCAUGUGGCAAUUAAAUGGUGGAGACACAGCAACAUCCACGGGCCAAA .......(((.(-(..((((((((((((--((.((....)).)))).(((((.....)))))))))))......(((....))).))..))...)).))).... ( -39.40, z-score = -1.49, R) >droSim1.chr2L 6068421 101 - 22036055 CUUUUUCGCCGG-UUUUGGCGUUGCCGC--CGUCGGCGACGUCGGCAACAUGUGUUGCAUGUGGCAAUUAAAUGGUGGAGACACAGCAACAUCCACGGGCCAAA .......(((.(-(..((((((((((((--((.((....)).)))).(((((.....)))))))))))......(((....))).))..))...)).))).... ( -39.40, z-score = -1.49, R) >droSec1.super_5 4341528 101 - 5866729 CUUUUUCGCCGG-UUUUGGCGUUGCCGC--CGUCGGCGACGUCGGCAACAUGUGUUGCAUGUGGCAAUUAAAUGGUGGAGACACAGCAACAUCCACGGGCCAAA .......(((.(-(..((((((((((((--((.((....)).)))).(((((.....)))))))))))......(((....))).))..))...)).))).... ( -39.40, z-score = -1.49, R) >droYak2.chr2L 15686787 102 + 22324452 CUUUUUCGCCGGUUUUUGGCGUUGCCGC--CGUCGGCGACGUCGGCAACAUGUGUUGCAUGUGGCAAUUAAAUGGUGGAGACACAGCAACAUCCACGGGCCAAA ..........((((..(((.(((((.((--((.((....)).))))..(((..(((((.....)))))...)))(((....))).)))))..)))..))))... ( -40.40, z-score = -1.80, R) >droEre2.scaffold_4929 15197507 101 - 26641161 CUUUUUCGCCGG-UUUUGGCAUUGCCGC--CGUCGGCGACGUCGGCAACAUGUGUUGCAUGUGGCAAUUAAAUGGUGGAGACACAGCAACAUCCACGGUCCAAA .......((((.-.....((((((((((--((.((....)).)))).(((((.....)))))))))))......(((....))).))........))))..... ( -37.84, z-score = -1.67, R) >dp4.chr4_group5 192735 97 + 2436548 CUCUUUUUUUG--UGUUGCUGCCGCCACAGCGGCGUUGGCGUCGGCAACAUGUGUUGCAUGUGGCAAUUAAAUGCCGG-----CAGCAACAGCAACGGAGCAAA ((((....(((--((((((((((((((((.(((((....)))))(((((....))))).))))))...........))-----)))))))).))).)))).... ( -42.80, z-score = -2.45, R) >droPer1.super_5 191705 94 + 6813705 ---CUUUUUUG--UGUUGCUGCCGCCACAGCGGCGUUGGCGUCGGCAACAUGUGUUGCAUGUGGCAAUUAAAUGCCGG-----CAGCAACAGCAACGGAGCAAA ---(((..(((--((((((((((((((((.(((((....)))))(((((....))))).))))))...........))-----)))))))).)))..))).... ( -43.60, z-score = -2.73, R) >droWil1.scaffold_180708 5202437 97 + 12563649 UUUUUUUAUCCAGCGUUGGCGUUGCCGU--UGCUGUUGUUGCCGGCAACAUGUGUUGCAUGUGGCAAUUAAAUGCCAG-----CAACAACACGAAGAAAGCAAA (((((((....((((.((((...)))).--))))((((((((..(((((....)))))...(((((......))))))-----)))))))..)))))))..... ( -34.90, z-score = -2.01, R) >droVir3.scaffold_12963 2918981 84 - 20206255 --------CU---UUUUUGCGCUGACGC--UGGCGUUGGCGUCGUCAACAUGUGUUGCAUGUGGCAAUUAAAUGCCAG-------GCAACACACAACAGCAAAA --------..---.((((((((..(((.--...)))..))..........((((((((...(((((......))))).-------)))))))).....)))))) ( -34.30, z-score = -2.92, R) >consensus CUUUUUCGCCGG_UUUUGGCGUUGCCGC__CGUCGGCGACGUCGGCAACAUGUGUUGCAUGUGGCAAUUAAAUGGUGGAGACACAGCAACAUCCACGGGCCAAA ........(((.....(((((((((((......)))))))))))((.(((((.....))))).))..............................)))...... (-21.46 = -21.24 + -0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:20:52 2011