
| Sequence ID | dm3.chr3RHet |
|---|---|
| Location | 2,273,935 – 2,274,053 |
| Length | 118 |
| Max. P | 0.617731 |

| Location | 2,273,935 – 2,274,053 |
|---|---|
| Length | 118 |
| Sequences | 7 |
| Columns | 130 |
| Reading direction | reverse |
| Mean pairwise identity | 61.27 |
| Shannon entropy | 0.75558 |
| G+C content | 0.56217 |
| Mean single sequence MFE | -40.20 |
| Consensus MFE | -12.42 |
| Energy contribution | -12.33 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.74 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.617731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
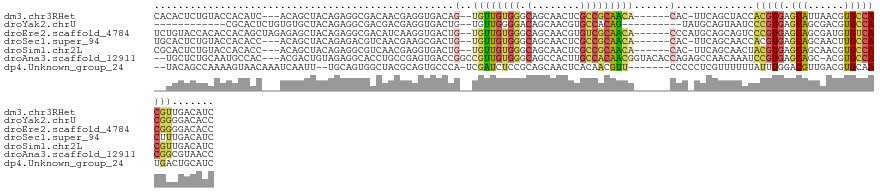
>dm3.chr3RHet 2273935 118 - 2517507 CACACUCUGUACCACAUC---ACAGCUACAGAGGCGACAACGAGGUGACAG--UGUUGUGGGCAGCAACUCGCCGCAACA------CAC-UUCAGCUACCACGUGAGCAUUAACGUGCCACGUUGACAUC ....(((((((.(.....---...).)))))))....(((((((.(((..(--((((((((.(........)))))))))------)..-.))).))..(((((........)))))...)))))..... ( -37.80, z-score = -1.51, R) >droYak2.chrU 4360173 106 + 28119190 ------------CGCACUCUGUGUGCUACAGAGGCGACGACGAGGUGACUG--UGUUGGGGACAGCAACGUGCCACAG----------UAUGCAGUAAUCCCGUGAGCAGCGACGUGCCACGGGGACACC ------------(((.(((((((...)))))))))).......((((((((--(((((....))))).(((((....)----------))))))))..(((((((.(((......)))))))))).)))) ( -45.50, z-score = -1.98, R) >droEre2.scaffold_4784 25241014 122 - 25762168 UCUGUACCACACCACAGCUAGAGAGCUACAGAGGCGACAUCAAGGUGACUG--UGUUGUGGGCAGCAACGUGUCGCAACA------CCCAUGCAGCAGUCCCGUGAGCAGCGAUGUGUCACGGGGACACC .((((.(((((.(((((.......(((.....)))..(((....))).)))--)).)))))))))....((((((((...------....)))......((((((((((....))).)))))))))))). ( -44.80, z-score = -0.72, R) >droSec1.super_94 86332 118 - 93855 UGCACUCUGUACCACACC---ACAGCUACAGAGACGUCAACGAAGCGACUG--UGUUGUGGGCAGCAACUCGCCGCAACA------CAC-UUCAGCAACCACGUGAGCAGCAACUUGCCACUUUGACAUC ....(((((((.(.....---...).)))))))..(((((.((((....((--((((((((.(........)))))))))------)))-))).........(((.((((....))))))).)))))... ( -37.40, z-score = -1.82, R) >droSim1.chr2L 950785 118 + 22036055 CGCACUCUGUACCACACC---ACAGCUACAGAGGCGUCAACGAGGUGACUG--UGUUGUGGGCAGCAACUCGCCGCAACA------CAC-UUCAGCAACUACGUGAGCAGCAACGUGCCACGUUGACAUC .((.(((((((.(.....---...).)))))))))((((((..(.(((.((--((((((((.(........)))))))))------)).-.))).)......(((.(((......))))))))))))... ( -45.20, z-score = -2.21, R) >droAna3.scaffold_12911 1886493 124 - 5364042 --UGCUCUGCAAUGCCAC---ACGACUGUAGAGGCACCUGCCGAGUGACCGGCCGUUGUGGGCAGCCACUUGCCACAACGGUACACCAGAGCCAACAAAUCCGUGAGCAGC-ACGUGCCACGGCGUAACC --.((((((((.((((.(---((....)).).))))..))).(.(((....((((((((((.(((....))))))))))))).)))))))))........(((((.(((..-...))))))))....... ( -45.10, z-score = -0.67, R) >dp4.Unknown_group_24 103240 118 + 109830 --UACAGCCAAAAGUAACAAAUCAAUU--UGCAGUGGCUACGCAGUGCCCA-UCGAUCUCCGCAGCAACUCACAACGUU-------CCCCCUCGUUUUUUAUUUGGACGUUGACGUGCAAUGACUGCAUC --...(((((...((((.........)--)))..)))))..(((((((...-.........)).(((.....(((((((-------(.................))))))))...)))....)))))... ( -25.63, z-score = -0.44, R) >consensus __CACUCUGUACCACACC___ACAGCUACAGAGGCGACAACGAGGUGACUG__UGUUGUGGGCAGCAACUCGCCGCAACA______CAC_UUCAGCAACCACGUGAGCAGCAACGUGCCACGGUGACAUC ............(((.........(((.....))).........)))......((((((((.(........)))))))))....................(((((.(((......))))))))....... (-12.42 = -12.33 + -0.09)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:49:07 2011