
| Sequence ID | dm3.chr3RHet |
|---|---|
| Location | 2,162,613 – 2,162,714 |
| Length | 101 |
| Max. P | 0.965195 |

| Location | 2,162,613 – 2,162,714 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 69.84 |
| Shannon entropy | 0.47961 |
| G+C content | 0.55337 |
| Mean single sequence MFE | -36.88 |
| Consensus MFE | -23.49 |
| Energy contribution | -24.36 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.965195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3RHet 2162613 101 + 2517507 ----GGCUGUAUUUUUGUAGCUUUGGCAUCUCCCGUUACAGCGCGGCGGCAGCGUGUGGUCGCUGUCGCCGAGUGUGGUUCGAAGGAUAGCACGGAGAAGAAGUG- ----(((((((....)))))))......((((((((....)))(((((((((((......))))))))))).((((...((....))..))))))))).......- ( -41.50, z-score = -2.27, R) >droWil1.scaffold_181075 650107 100 - 1141854 ----GGCGGCGCAUUUGU-GGUUUAAGAAGAACCGUUUUAGCGGGGCGCCAGCGUUCUAUCGCUGUCGCCGAGUGUGGUUCGGUGGAAGGCGCGCACAAGAAUUG- ----.((..(((.....(-(((((.....)))))).....)))..(((((((((......)))).((((((((.....))))))))..)))))))..........- ( -36.00, z-score = -0.45, R) >droAna3.scaffold_13111 1420772 105 + 1527837 CUGUGUGUGUGUGUUGUC-GCUUUAAGAAAUGUCGUUUUAGCAGGGCGACAGUGUAGUGUCGCUGUCGCCGAGUGUGGUUCGGUGGUAAGGAUAGAAAGAAGCAGG ((((.....((..(((((-(((((...(((......)))...))))))))))..)).((((..(..(((((((.....)))))))..)..)))).......)))). ( -32.30, z-score = -2.69, R) >droSim1.chrU 11369358 101 - 15797150 ----GGCUGUAUUUUUGUAGCUUUAGCAUCUCCCGUUUCAGCGCGGCGGCAGUGUGUGUUCGCUGUCGCCGAGUGUGGCUCGAAGGUUAGCACGGAGAAGAAGUG- ----.((((.(((((((.((((...(((.(((.(((....))).((((((((((......)))))))))))))))))))))))))))))))..............- ( -37.70, z-score = -1.55, R) >consensus ____GGCUGUAUUUUUGU_GCUUUAACAACUCCCGUUUCAGCGCGGCGGCAGCGUGUGGUCGCUGUCGCCGAGUGUGGUUCGAAGGAAAGCACGGAGAAGAAGUG_ ............((((....((.......((((((.....((.(((((((((((......))))))))))).))......)))))).......))....))))... (-23.49 = -24.36 + 0.88)


| Location | 2,162,613 – 2,162,714 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 69.84 |
| Shannon entropy | 0.47961 |
| G+C content | 0.55337 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -14.09 |
| Energy contribution | -14.28 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.938750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
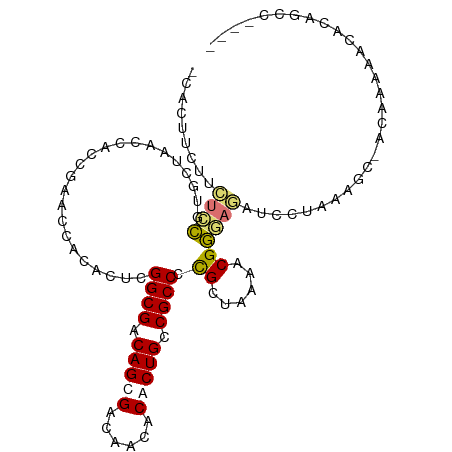
>dm3.chr3RHet 2162613 101 - 2517507 -CACUUCUUCUCCGUGCUAUCCUUCGAACCACACUCGGCGACAGCGACCACACGCUGCCGCCGCGCUGUAACGGGAGAUGCCAAAGCUACAAAAAUACAGCC---- -.......((((((((...((....))..))).(.(((((.(((((......))))).))))).)........))))).......(((..........))).---- ( -26.60, z-score = -1.34, R) >droWil1.scaffold_181075 650107 100 + 1141854 -CAAUUCUUGUGCGCGCCUUCCACCGAACCACACUCGGCGACAGCGAUAGAACGCUGGCGCCCCGCUAAAACGGUUCUUCUUAAACC-ACAAAUGCGCCGCC---- -........(((.((((...................((((.(((((......))))).))))..........((((.......))))-......))))))).---- ( -27.80, z-score = -1.48, R) >droAna3.scaffold_13111 1420772 105 - 1527837 CCUGCUUCUUUCUAUCCUUACCACCGAACCACACUCGGCGACAGCGACACUACACUGUCGCCCUGCUAAAACGACAUUUCUUAAAGC-GACAACACACACACACAG ..(((((..................((.......))((((((((.(......).)))))))).....................))))-)................. ( -17.60, z-score = -3.19, R) >droSim1.chrU 11369358 101 + 15797150 -CACUUCUUCUCCGUGCUAACCUUCGAGCCACACUCGGCGACAGCGAACACACACUGCCGCCGCGCUGAAACGGGAGAUGCUAAAGCUACAAAAAUACAGCC---- -..(((((..((.((((........(((.....)))((((.(((.(......).))).)))))))).))...)))))..((....))...............---- ( -24.80, z-score = -1.02, R) >consensus _CACUUCUUCUCCGUGCUAACCACCGAACCACACUCGGCGACAGCGACAACACACUGCCGCCCCGCUAAAACGGGAGAUCCUAAAGC_ACAAAAACACAGCC____ .........((((.......................((((.(((((......))))).)))).((......))))))............................. (-14.09 = -14.28 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:49:05 2011