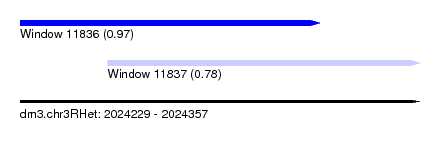
| Sequence ID | dm3.chr3RHet |
|---|---|
| Location | 2,024,229 – 2,024,357 |
| Length | 128 |
| Max. P | 0.965395 |

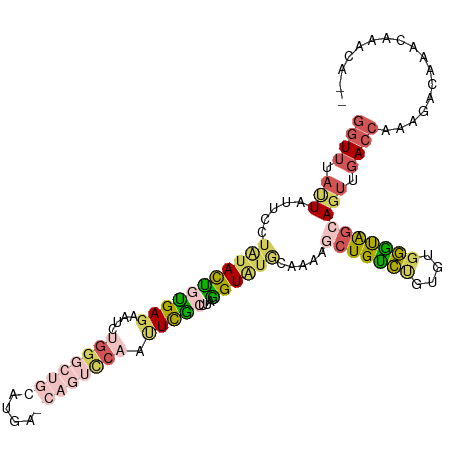
| Location | 2,024,229 – 2,024,325 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 64.00 |
| Shannon entropy | 0.63652 |
| G+C content | 0.43781 |
| Mean single sequence MFE | -28.28 |
| Consensus MFE | -17.10 |
| Energy contribution | -18.90 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.965395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3RHet 2024229 96 + 2517507 GGUUUAUUAUUCCUGUACUGUGAGAGUCUGG-------------UUCAAUUCGCUUAGGUAUACAAAAGCUGUCUGUGUGGGUAGCAGUUGACCAAAGACAAACAAACA-- .((((....((((......).))).((((((-------------(.(((((.(((....((((((.........))))))...)))))))))))..)))))))).....-- ( -17.90, z-score = 1.27, R) >droSim1.chr2h_random 2114562 109 - 3178526 GGUUUAUUAUUCCUACACUCUGAGAAUCCGGGCUGCAUGAGCAGUCCAACUCGCUUAGGUAUGCAAAACCUGCCUGUGUGGGUGGCAGUUGACCAAAGACAGACCAACA-- (((((......(((((((((((((.....(((((((....)))))))......))))))...(((.....)))..)))))))((((....).))).....)))))....-- ( -35.10, z-score = -1.38, R) >droSec1.super_83 84317 111 - 109685 GGUUUACUAUUCCUAUACCGUGAGAAUGUGGGCUGCAUGAGCAGUCCAAUUCACUUGGGUAUGCAAAAGCUGCCUGUGUGGGUAGCAGUUGACCAAAGACAAACAACAAAA ((((.(((.(((((((((.(((((....((((((((....)))))))).)))))..(((((.((....)))))))))))))).)).))).))))................. ( -40.80, z-score = -3.34, R) >droYak2.chrU 7471682 109 + 28119190 GGUGUACUAUUCCUAUACUGUGAGAAACUGGGCUGCAUGAACAGUCCAAUUCGCUUAGGUAUGAAAAAGCUGUCCGUGUGGGCAGCAGUUGACCGAAGCCAAACAAACA-- (((..(((...((((....(((((....(((((((......))))))).))))).)))).........(((((((....))))))))))..)))...............-- ( -33.70, z-score = -1.71, R) >droAna3.scaffold_13417 3121590 84 - 6960332 --UUUCUUACGCUUCUAUAUCGAUGGAAGGGA------------------UUGCUCAUGUG-----GAGCUGUAGCUAUGUAUAAAACCCGAAACAAAA-AAACAAAUUC- --((((...(.(((((((....))))))).).------------------..((((.....-----))))....................)))).....-..........- ( -13.90, z-score = -0.35, R) >consensus GGUUUAUUAUUCCUAUACUGUGAGAAUCUGGGCUGCAUGA_CAGUCCAAUUCGCUUAGGUAUGCAAAAGCUGUCUGUGUGGGUAGCAGUUGACCAAAGACAAACAAACA__ ((((.(((.....(((((((((((....((((((((....)))))))).)))))...)))))).....(((((((....)))))))))).))))................. (-17.10 = -18.90 + 1.80)

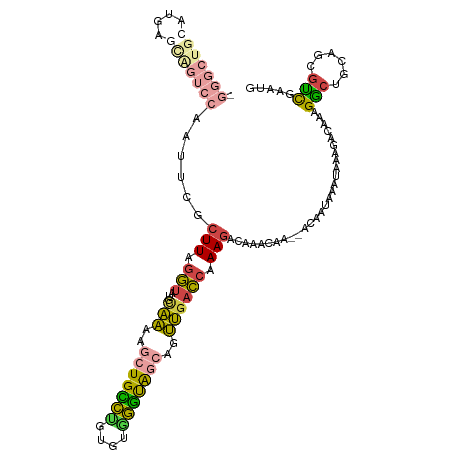
| Location | 2,024,257 – 2,024,357 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 68.75 |
| Shannon entropy | 0.58915 |
| G+C content | 0.45232 |
| Mean single sequence MFE | -28.13 |
| Consensus MFE | -15.40 |
| Energy contribution | -15.77 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.62 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.782428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3RHet 2024257 100 + 2517507 -------------UGGUUCAAUUCGCUUAGGUAUACAAAAGCUGUCUGUGUGGGUAGCAGUUGACCAAAGACAAACAA--ACAAUAAAUGAAUACAAAGGCUGCAGCGUCGAAUG -------------..((((.((((((((..(....)..))))(((((...(((...........))).))))).....--.........)))).....(((......))))))). ( -18.30, z-score = 0.75, R) >droSim1.chr2h_random 2114590 113 - 3178526 CGGGCUGCAUGAGCAGUCCAACUCGCUUAGGUAUGCAAAACCUGCCUGUGUGGGUGGCAGUUGACCAAAGACAGACCA--ACAAUAAAUAUAGACAAAGGAUGCAGCGUCGAAUG ((((((((((((((((.....)).)))).(((.(((...(((..(....)..))).)))....)))............--....................))))))).))).... ( -31.70, z-score = -0.53, R) >droSec1.super_83 84345 115 - 109685 UGGGCUGCAUGAGCAGUCCAAUUCACUUGGGUAUGCAAAAGCUGCCUGUGUGGGUAGCAGUUGACCAAAGACAAACAACAAAAAAAAAUAAAGACAAAGACUGCAGCGUCGAAUG ((((((((....))))))))((((.(((.(((...(((..(((((((....)))))))..)))))).)))............................(((......))))))). ( -35.00, z-score = -2.40, R) >droYak2.chrU 7471710 113 + 28119190 UGGGCUGCAUGAACAGUCCAAUUCGCUUAGGUAUGAAAAAGCUGUCCGUGUGGGCAGCAGUUGACCGAAGCCAAACAA--ACAACAACUAAAGACAAAGGCUGCAGCGUCGAAUG (((((((......)))))))((((((((.(((....((..(((((((....)))))))..)).))).)))).......--............(((....((....))))))))). ( -34.20, z-score = -1.49, R) >droEre2.scaffold_4929 25167561 113 + 26641161 CGGGCUGCAUGAGUGGUCCAAUUCGCUUAGGUAUGCAAAAGGUGCCUGUGUAGGUAGCAGUUGACCAAAGACGAACAA--ACAAUAAAUAAAGACAAAGGCUGCAGCGUCGAAUG ((((((((((((((((......)))))))(((...(((..(.(((((....))))).)..))))))............--.....................)))))).))).... ( -28.50, z-score = -0.33, R) >droAna3.scaffold_13417 3121618 88 - 6960332 ------------------------GAUUGCUCAUGUGGA-GCUGUAGCUAUGUAUAAAACCCGAAACAAAA-AAACAA-AUUCAUGCUUGAAUUGCAUGCCCGCAGAGGUGAUUG ------------------------(((..((..((((((-((....)))......................-......-...(((((.......))))).)))))..))..))). ( -21.10, z-score = -1.68, R) >consensus _GGGCUGCAUGAGCAGUCCAAUUCGCUUAGGUAUGCAAAAGCUGCCUGUGUGGGUAGCAGUUGACCAAAGACAAACAA__ACAAUAAAUAAAGACAAAGGCUGCAGCGUCGAAUG .(((((((....)))))))......(((.(((...(((..(((((((....)))))))..)))))).)))............................(((......)))..... (-15.40 = -15.77 + 0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:48:59 2011