| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,260,957 – 6,261,057 |
| Length | 100 |
| Max. P | 0.957981 |

| Location | 6,260,957 – 6,261,057 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 78.12 |
| Shannon entropy | 0.46027 |
| G+C content | 0.41085 |
| Mean single sequence MFE | -27.27 |
| Consensus MFE | -19.93 |
| Energy contribution | -19.93 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.957981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
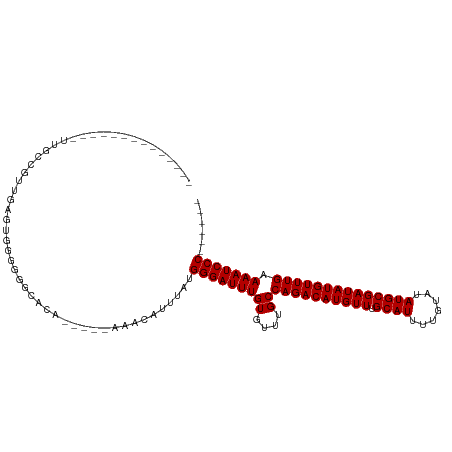
>dm3.chr2L 6260957 100 - 23011544 --UUCCCUUCGUG---GUUGUUGGGUGGGGGGCACA-----AAACAUUUAUGGGAUUUGUGUUUGCCAGACAUGUUUGCAUUUUGUAUAUGCGAUAUGUUUGAAAAUCCC------ --..(((..((..---..))..))).((((((((((-----((.(........).)))))))))..((((((((((.((((.......)))))))))))))).....)))------ ( -27.60, z-score = -0.67, R) >droSim1.chr2L 6058909 103 - 22036055 --UUCCCUUCGUGGCACUCGUUGGGUGGGGGGCACA-----AAACAUUUAUGGGAUUUGUGUUUGCCAGACAUGUUUGCAUUUUGUAUAUGCGAUAUGUUUGAAAAUCCC------ --.(((((.(.((((....)))).).))))).....-----..........(((((((((....))((((((((((.((((.......)))))))))))))).)))))))------ ( -29.00, z-score = -0.64, R) >droSec1.super_5 4332046 103 - 5866729 --UUCCCUUCGUGGUAGUCGUUGGGUGGGGGGCACA-----AAACAUUUAUGGGAUUUGUGUUUGCCAGACAUGUUUGCAUUUUGUAUAUGCGAUAUGUUUGAAAAUCCC------ --..(((..((.......))..))).((((((((((-----((.(........).)))))))))..((((((((((.((((.......)))))))))))))).....)))------ ( -28.70, z-score = -0.86, R) >droYak2.chr2L 15677103 91 + 22324452 --------------UUCCCUUCCAGGGGGGGGCACA-----AAACAUUUAUGGGAUUUGUGUUUGCCAGACAUGUUUGCAUUUUGUAUAUGCGAUAUGUUUGAAAAUCCC------ --------------..((((((....))))))....-----..........(((((((((....))((((((((((.((((.......)))))))))))))).)))))))------ ( -28.30, z-score = -1.64, R) >droEre2.scaffold_4929 15187996 89 - 26641161 --------------UUCCCUUCCA--GGGGGGCACA-----AAACAUUUAUGGGAUUUGUGUUUGCCAGACAUGUUUGCAUUUUGUAUAUGCGAUAUGUUUGAAAAUCCC------ --------------..(((((...--.)))))....-----..........(((((((((....))((((((((((.((((.......)))))))))))))).)))))))------ ( -25.60, z-score = -1.42, R) >droAna3.scaffold_12916 8223970 94 + 16180835 -----------CCCCUAACACCCACGAUGGGGCACA-----AAACAUUUAUGGGAUUUGUGUUUGCCAGACAUGUUUGCAUUUUGUAUAUGCGAUAUGUUUGAAAAUCCC------ -----------((((.............))))....-----..........(((((((((....))((((((((((.((((.......)))))))))))))).)))))))------ ( -25.32, z-score = -1.44, R) >dp4.chr4_group5 175147 97 + 2436548 --------------UUCCCAUUGAGAGAGGGGCACA-----AAACAUUUAUGGGAUUUGUGUUUGCCAGACAUGUUUGCAUUUUGUAUAUGCGAUAUGUUUGAAAAUCCCCUCCGU --------------..........(.((((((((((-----((.(........).)))))).....((((((((((.((((.......)))))))))))))).....))))))).. ( -29.00, z-score = -2.41, R) >droPer1.super_5 174975 96 + 6813705 ---------------UUCCAUUGAGAGAGGGGCACA-----AAACAUUUAUGGGAUUUGUGUUUGCCAGACAUGUUUGCAUUUUGUAUAUGCGAUAUGUUUGAAAAUCCCCUCCGU ---------------.........(.((((((((((-----((.(........).)))))).....((((((((((.((((.......)))))))))))))).....))))))).. ( -29.00, z-score = -2.49, R) >droWil1.scaffold_180708 5185862 105 + 12563649 CUUUCCAUUUUGCCAUUUUUCUUAUUUUAUCACACA-----AAACAUUUAUGGGAUUUGUGUUUGCCAGACAUGUUUGCAUUUUGUAUAUGCGAUAUGUUUGAAAAUCCC------ ....................................-----..........(((((((((....))((((((((((.((((.......)))))))))))))).)))))))------ ( -19.80, z-score = -1.36, R) >droVir3.scaffold_12963 17274790 97 + 20206255 ------ACCCAGUCCAGCACCUCA--GCACCACACA-----AAACAUUUAUGGGAUUUGUGUUUGCCAGACAUGUUUGCAUUUUGUAUAUGCGAUAUGUUUGAAAAUCCC------ ------..................--(((..(((((-----((.(........).))))))).)))((((((((((.((((.......))))))))))))))........------ ( -21.50, z-score = -0.95, R) >droMoj3.scaffold_6500 19126150 106 + 32352404 ----GUCUCGAAAAGGGAGGCGGGGACGCAGACACACACACAGACAUUUAUGGGAUUUGUGUUUGCCAGACAUGUUUGCAUUUUGUAUAUGCGAUAUGUUUGAAAAUCCC------ ----(((((.......))))).((((.(((((((((....((........)).....)))))))))((((((((((.((((.......))))))))))))))....))))------ ( -36.40, z-score = -3.81, R) >droGri2.scaffold_15252 14646165 102 + 17193109 ---CACGGCUCAGAGGGAGGCAGGUUGGGGGACUCA-----AAACAUUUAUGGGAUUUGUGUUUGCCAGACAUGUUUGCAUUUUGUAUAUGCGAUAUGUUUGAAAAUCCC------ ---...(.(((.....))).)...((((....).))-----).........(((((((((....))((((((((((.((((.......)))))))))))))).)))))))------ ( -27.00, z-score = -0.81, R) >consensus ______________UUGCCGUUGAGUGGGGGGCACA_____AAACAUUUAUGGGAUUUGUGUUUGCCAGACAUGUUUGCAUUUUGUAUAUGCGAUAUGUUUGAAAAUCCC______ ...................................................(((((((((....))((((((((((.((((.......)))))))))))))).)))))))...... (-19.93 = -19.93 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:20:51 2011