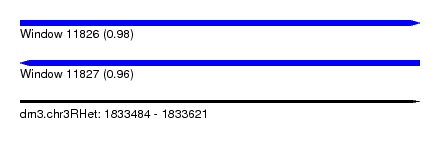
| Sequence ID | dm3.chr3RHet |
|---|---|
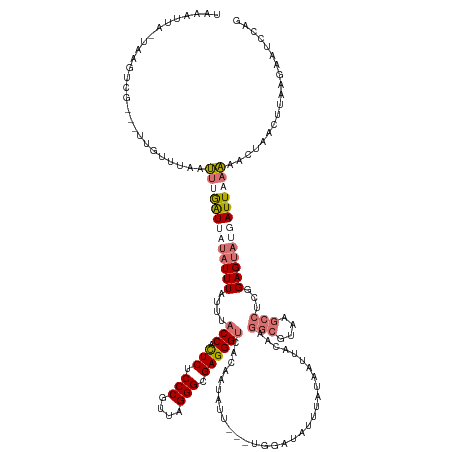
| Location | 1,833,484 – 1,833,621 |
| Length | 137 |
| Max. P | 0.979060 |

| Location | 1,833,484 – 1,833,621 |
|---|---|
| Length | 137 |
| Sequences | 4 |
| Columns | 144 |
| Reading direction | forward |
| Mean pairwise identity | 68.99 |
| Shannon entropy | 0.50300 |
| G+C content | 0.35072 |
| Mean single sequence MFE | -29.62 |
| Consensus MFE | -13.95 |
| Energy contribution | -18.45 |
| Covariance contribution | 4.50 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.979060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3RHet 1833484 137 + 2517507 CCGGAUUCUUAAGUUAGUUUUUAAUCAUACUCCGAGGCUUACGCCUUGUAAUUAUAAAUACCCA---AAUAUUGUGACCCUCGCCCUAACGGGAGAGUGGUAAAUAAAUAUCAUUAAAUUAACCAA---CGACUUA-UAAUUUC ..(((((..((((((.(((((((((.(((...((((((....))))))................---..((((...((((((.(((....))).))).))).))))..))).))))))......))---)))))))-.))))). ( -29.70, z-score = -2.13, R) >droSec1.super_59 92262 136 + 165940 CUGAAUUCUUAAGUUAGUUUUUAAUCAUACUCCGAGGCUUACGCCUUGUAAUUAUAAAUACCCA---AAUAUUGUGACCCUCGCCCUAACGGGAGAGUGGUAAAUAAAUAUAACCAAACUAAACAAA--CGACUUA-UAAUU-- .........((((((.((((............((((((....))))))................---.(((((...((((((.(((....))).))).))).....))))).............)))--)))))))-.....-- ( -29.60, z-score = -3.05, R) >droYak2.chr2h 314306 139 - 894407 UUAGAUUUUUAA-UCAGUUAUAAAUCAUACUCCGUGGCUUACGCCUCGUA-UUGUAAACCUACAUAAAAAAUUAAGACCCUCGCCCUAACGGGAGAGUGGUAA-UAAAUA-AAUCAACUUAAAACACCACGACUUA-UAAUUUA ...(((.....)-))((((((((.((.(((..((.(((....))).))..-..)))...............(((..((((((.(((....))).))).)))..-)))...-...................)).)))-))))).. ( -27.70, z-score = -2.54, R) >droEre2.scaffold_4784 24528783 120 + 25762168 --------UUAAGUUAGUCUAAAAUUGUACUCCGGGGUUUA------ACAGCCUCGAUAAUCUG-----AAUUAA-ACCCUCGCCCUAACGGGAGAACGGGGAAUAAA-GUUAUCAAAUUAUUAAC---CAACCGAGUGAUUUA --------.(((((((.((.....((((.((((((((((((------((((..........)))-----..))))-))))((.(((....))).)).))))).)))).-((((.........))))---.....)).))))))) ( -31.50, z-score = -1.91, R) >consensus CUGGAUUCUUAAGUUAGUUUUAAAUCAUACUCCGAGGCUUACGCCUUGUAAUUAUAAAUACCCA___AAAAUUAAGACCCUCGCCCUAACGGGAGAGUGGUAAAUAAAUAUAAUCAAAUUAAAAAA___CGACUUA_UAAUUUA .........((((((.................((((((....))))))............................((((((.(((....))).))).))).............................))))))........ (-13.95 = -18.45 + 4.50)

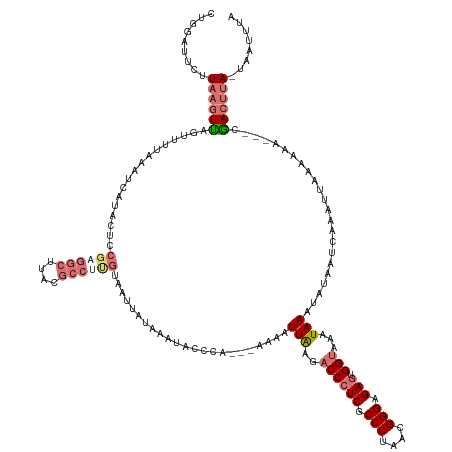
| Location | 1,833,484 – 1,833,621 |
|---|---|
| Length | 137 |
| Sequences | 4 |
| Columns | 144 |
| Reading direction | reverse |
| Mean pairwise identity | 68.99 |
| Shannon entropy | 0.50300 |
| G+C content | 0.35072 |
| Mean single sequence MFE | -33.05 |
| Consensus MFE | -18.44 |
| Energy contribution | -20.75 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.958967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3RHet 1833484 137 - 2517507 GAAAUUA-UAAGUCG---UUGGUUAAUUUAAUGAUAUUUAUUUACCACUCUCCCGUUAGGGCGAGGGUCACAAUAUU---UGGGUAUUUAUAAUUACAAGGCGUAAGCCUCGGAGUAUGAUUAAAAACUAACUUAAGAAUCCGG .......-......(---((((((..((((((.((((((....(((.(((.(((....))).)))))).........---................(.((((....)))).))))))).)))))))))))))............ ( -34.80, z-score = -2.13, R) >droSec1.super_59 92262 136 - 165940 --AAUUA-UAAGUCG--UUUGUUUAGUUUGGUUAUAUUUAUUUACCACUCUCCCGUUAGGGCGAGGGUCACAAUAUU---UGGGUAUUUAUAAUUACAAGGCGUAAGCCUCGGAGUAUGAUUAAAAACUAACUUAAGAAUUCAG --.....-....((.--...(((...(((((((((((((....(((.(((.(((....))).)))))).........---................(.((((....)))).))))))))))))))....)))....))...... ( -32.50, z-score = -1.94, R) >droYak2.chr2h 314306 139 + 894407 UAAAUUA-UAAGUCGUGGUGUUUUAAGUUGAUU-UAUUUA-UUACCACUCUCCCGUUAGGGCGAGGGUCUUAAUUUUUUAUGUAGGUUUACAA-UACGAGGCGUAAGCCACGGAGUAUGAUUUAUAACUGA-UUAAAAAUCUAA ....(((-(((((((((.(.......((.((((-((((((-..(((.(((.(((....))).))))))..)))........))))))).))..-..((.(((....))).)).).))))))))))))....-............ ( -36.20, z-score = -2.51, R) >droEre2.scaffold_4784 24528783 120 - 25762168 UAAAUCACUCGGUUG---GUUAAUAAUUUGAUAAC-UUUAUUCCCCGUUCUCCCGUUAGGGCGAGGGU-UUAAUU-----CAGAUUAUCGAGGCUGU------UAAACCCCGGAGUACAAUUUUAGACUAACUUAA-------- ......((((.(..(---(((((((.(((((((((-((((..(((((((((......)))))).))).-.)))..-----.)).))))))))..)))------).)))).).))))....................-------- ( -28.70, z-score = -0.77, R) >consensus UAAAUUA_UAAGUCG___UUGUUUAAUUUGAUUAUAUUUAUUUACCACUCUCCCGUUAGGGCGAGGGUCACAAUAUU___UGGAUAUUUAUAAUUACAAGGCGUAAGCCUCGGAGUAUGAUUAAAAACUAACUUAAGAAUCCAG ..........................(((((((((((((....(((.(((.(((....))).))))))...............................(((....)))...)))))))))))))................... (-18.44 = -20.75 + 2.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:48:51 2011