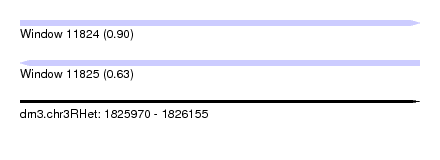
| Sequence ID | dm3.chr3RHet |
|---|---|
| Location | 1,825,970 – 1,826,155 |
| Length | 185 |
| Max. P | 0.899622 |

| Location | 1,825,970 – 1,826,155 |
|---|---|
| Length | 185 |
| Sequences | 4 |
| Columns | 193 |
| Reading direction | forward |
| Mean pairwise identity | 54.76 |
| Shannon entropy | 0.75314 |
| G+C content | 0.56808 |
| Mean single sequence MFE | -73.03 |
| Consensus MFE | -21.33 |
| Energy contribution | -26.20 |
| Covariance contribution | 4.87 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.899622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
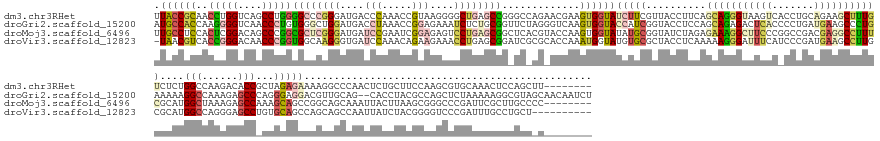
>dm3.chr3RHet 1825970 185 + 2517507 UUACCGCAACCUGGUCAGCCUGGGGCCCGGGAUGACCCAAACCGUAAGGGGCUGAGCCGGGCCAGAACGAAGUGGUAUCUUCGUUACCUUCAGCAGGGUAAGUCACCUGCAGAAGCUUUGUCUCUGGCCAAGACACCGCUAGAGAAAAGGCCCAACUCUGCUUCCAAGCGUGCAAACUCCAGCUU-------- .....(((....(((((.(((((...))))).))))).....(((..(((((.(((..(((((..(((((((......)))))))..((((.(((((........))))).)))).....((((((((.........))))))))...)))))..))).)).)))..))))))............-------- ( -71.40, z-score = -1.45, R) >droGri2.scaffold_15200 144791 191 + 219039 AUGCCACCAAGGGGUCAACCCUGUGGGCUGGAUGACCUAAACCGGAGAAAUCUCUGCGGUUCUAGGGUCAAGUGGUACCAUCGGUACCUCCAGCAGAGACUCACCCCUGAUGAAGCCCUGAAAAAGGCCAAAGAGCCCAGGGAGGACGUUGCAG--CACCUACGCCAGCUCUAAAAAGGCGUAGCAACAAUCU .(((..(((.((((....)))).)))((((((((((((.(((((.(((....))).)))))...))))))...((((((...)))))))))))).(..((((..(((((.....(((.(....).))).........)))))..)).))..).)--)).(((((((...........)))))))......... ( -73.74, z-score = -2.37, R) >droMoj3.scaffold_6496 25219154 185 - 26866924 UUGCCUCCACUCGGACAGCCCGGCGCUCGGGAUGAUCCGAAUCGGAGAGUCCUGAGCGGCUCACGUACCAAGUGGUAUAUGCGGUAUCUAGAGAAAGGCUUCCCGGCCGACGAGGCCUUUCGCAUGGCUAAAGAGCCAAAGCAGCCGGCAGCAAAUUACUUAAGCGGGCCCGAUUCGCUUGCCCC-------- ((((.(((....)))..(((.((((((((((((..((((...))))..)))))))))(((((..(((((....)))))(((((....)....((((((((((.........)))))))))))))).......)))))......)))))).)))).........((((((.......))))))...-------- ( -76.60, z-score = -1.86, R) >droVir3.scaffold_12823 181990 182 - 2474545 -UAACGUCACCGGGACAACCCGGUGGCAAGGGUGAUCCAAACAGAAGAAACCUGAGCGGAUCGCGCACCAAAUGGUAUGUGCGCUACCUCAAAAAGGGAUUUCAUCCCGAUGAAGCCUUGCGCAUGGCCAGGGAGCCUGUGCAGCCAGCAGCCAAUUAUCUACGGGGUCCCGAUUUGCCUGCU---------- -......(((((((....)))))))((((((.((((((...(((.......)))...)))))((((((..........))))))....(((....(((((...)))))..)))).))))))(((.(((..((((.(((((((.....))((........))))))).)))).....)))))).---------- ( -70.40, z-score = -1.91, R) >consensus UUACCGCCACCGGGACAACCCGGUGCCCGGGAUGACCCAAACCGGAGAAACCUGAGCGGAUCAAGAACCAAGUGGUAUCUGCGGUACCUCCAGAAGGGAUUCCCACCCGAUGAAGCCUUGCGCAUGGCCAAAGAGCCAAUGCAGCAAGCAGCAAAUUACCUACGCGAGCCCGAAAAGCCCGCC_C________ .((((((..(((((....)))))((((((((....(((.....)))....)))))))).............))))))(((((..........(((((((((((.......)))))))))))....(((......)))...)))))................................................ (-21.33 = -26.20 + 4.87)
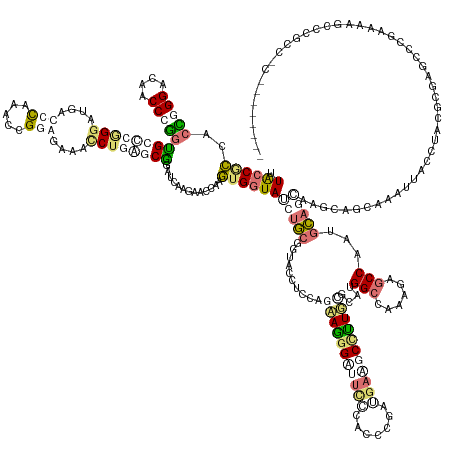
| Location | 1,825,970 – 1,826,155 |
|---|---|
| Length | 185 |
| Sequences | 4 |
| Columns | 193 |
| Reading direction | reverse |
| Mean pairwise identity | 54.76 |
| Shannon entropy | 0.75314 |
| G+C content | 0.56808 |
| Mean single sequence MFE | -76.65 |
| Consensus MFE | -20.91 |
| Energy contribution | -22.16 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.75 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.633514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3RHet 1825970 185 - 2517507 --------AAGCUGGAGUUUGCACGCUUGGAAGCAGAGUUGGGCCUUUUCUCUAGCGGUGUCUUGGCCAGAGACAAAGCUUCUGCAGGUGACUUACCCUGCUGAAGGUAACGAAGAUACCACUUCGUUCUGGCCCGGCUCAGCCCCUUACGGUUUGGGUCAUCCCGGGCCCCAGGCUGACCAGGUUGCGGUAA --------..((((.((..((((.((((((..((.((((((((((.........((..((((((.....))))))..))(((.(((((........))))).)))((.(((((((......))))))))))))))))))).)).......((((((((....))))))))))))))))..))..)).)))).. ( -78.40, z-score = -1.79, R) >droGri2.scaffold_15200 144791 191 - 219039 AGAUUGUUGCUACGCCUUUUUAGAGCUGGCGUAGGUG--CUGCAACGUCCUCCCUGGGCUCUUUGGCCUUUUUCAGGGCUUCAUCAGGGGUGAGUCUCUGCUGGAGGUACCGAUGGUACCACUUGACCCUAGAACCGCAGAGAUUUCUCCGGUUUAGGUCAUCCAGCCCACAGGGUUGACCCCUUGGUGGCAU .........(((((((...........)))))))(((--(..(...(((((....(((((....))))).....)))))......((((((.((.((((((((((((((((...))))))...(((((...((((((..(((....))))))))).)))))))))))....)))))).))))))..)..)))) ( -80.90, z-score = -2.55, R) >droMoj3.scaffold_6496 25219154 185 + 26866924 --------GGGGCAAGCGAAUCGGGCCCGCUUAAGUAAUUUGCUGCCGGCUGCUUUGGCUCUUUAGCCAUGCGAAAGGCCUCGUCGGCCGGGAAGCCUUUCUCUAGAUACCGCAUAUACCACUUGGUACGUGAGCCGCUCAGGACUCUCCGAUUCGGAUCAUCCCGAGCGCCGGGCUGUCCGAGUGGAGGCAA --------.((((..(.....)..))))((............(((.(((((((..(((((....))))).))(((((((((((.....))))..)))))))...........((..((((....))))..)))))))..)))....((((.(((((((.((.((((.....)))).))))))))))))))).. ( -71.70, z-score = 0.24, R) >droVir3.scaffold_12823 181990 182 + 2474545 ----------AGCAGGCAAAUCGGGACCCCGUAGAUAAUUGGCUGCUGGCUGCACAGGCUCCCUGGCCAUGCGCAAGGCUUCAUCGGGAUGAAAUCCCUUUUUGAGGUAGCGCACAUACCAUUUGGUGCGCGAUCCGCUCAGGUUUCUUCUGUUUGGAUCACCCUUGCCACCGGGUUGUCCCGGUGACGUUA- ----------(((((.(((..(((....))).......))).))))).((.((.((((...))))))...))(((((((((((..(((((...)))))....)))))..((((((..........))))))((((((..((((.....))))..))))))..))))))(((((((....)))))))......- ( -75.60, z-score = -2.37, R) >consensus ________G_GGCAGGCUAAUCGGGCCCGCGUAGAUAAUUGGCUGCUGGCUCCAUAGGCUCCUUGGCCAUGCGCAAGGCUUCAUCAGGAGGGAAUCCCUGCUGGAGGUACCGAAGAUACCACUUGGUACGAGAACCGCUCAGGUCUCUCCGGUUUGGAUCAUCCCGGCCACCGGGCUGACCCGGUGGCGGCAA ......................((((((((((((........))))))).......)))))....(((.........((((((..((((.....))))....))))))..(((.(((........))).)))..((((((.((((..((((((((((......))))..))))))..)))))))))).))).. (-20.91 = -22.16 + 1.25)
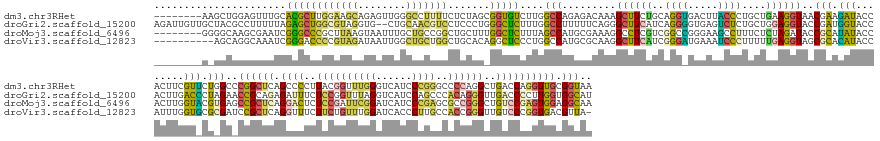
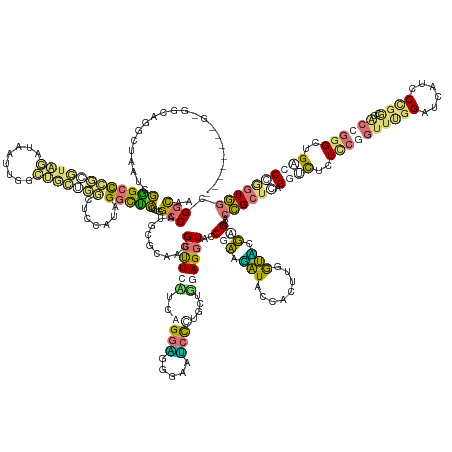
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:48:49 2011