
| Sequence ID | dm3.chr3RHet |
|---|---|
| Location | 1,746,143 – 1,746,200 |
| Length | 57 |
| Max. P | 0.844288 |

| Location | 1,746,143 – 1,746,200 |
|---|---|
| Length | 57 |
| Sequences | 5 |
| Columns | 58 |
| Reading direction | forward |
| Mean pairwise identity | 59.82 |
| Shannon entropy | 0.71435 |
| G+C content | 0.45044 |
| Mean single sequence MFE | -11.12 |
| Consensus MFE | -5.18 |
| Energy contribution | -5.34 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.60 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.844288 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
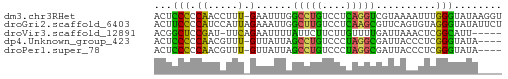
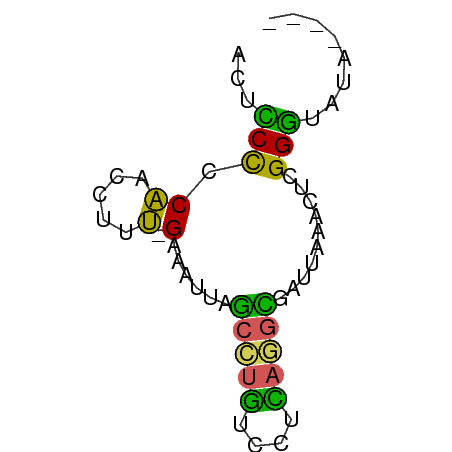
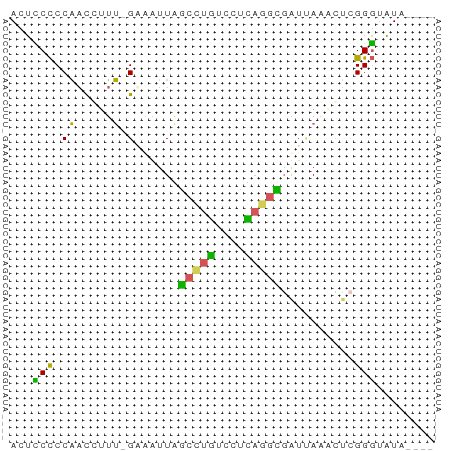
>dm3.chr3RHet 1746143 57 + 2517507 ACUCCCCCAACCUUU-GAAUUUGGCCUGUCCUCAGGUCGUAAAAUUUGGGUAUAAGGU .....(((((..(((-(....(((((((....)))))))))))..)))))........ ( -15.20, z-score = -1.27, R) >droGri2.scaffold_6403 3524 58 + 10120 ACUUCCCCAUCCAUUAGAAAUUGGCUUGUCCUCAAGCGUUCAGUGUAGGGUAUAUUCU .....(((...((((.(((....(((((....))))).)))))))..)))........ ( -12.40, z-score = -1.52, R) >droVir3.scaffold_12891 417275 52 + 459118 ACGGCUCCGAU-UUCAGAAUUUUAUUCUUCUUGUUUUGAUUAAACUCGGCAUU----- ......((((.-.(((((((............)))))))......))))....----- ( -6.40, z-score = -0.37, R) >dp4.Unknown_group_423 39570 53 + 54704 ACUCCCCCAACGUUU-GUUAUUAGCCUGUCCCUAGGCGAUUACCCUCGGGUAUA---- ...............-.......(((((....)))))...((((....))))..---- ( -10.80, z-score = -0.61, R) >droPer1.super_78 178794 53 - 255460 ACUCCCCCAACGUUU-GUUAUUAGCCUGUCCCUAGGCGAUUACCCUCGGGUAUA---- ...............-.......(((((....)))))...((((....))))..---- ( -10.80, z-score = -0.61, R) >consensus ACUCCCCCAACCUUU_GAAAUUAGCCUGUCCUCAGGCGAUUAAACUCGGGUAUA____ .....(((...............(((((....)))))((......)))))........ ( -5.18 = -5.34 + 0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:48:45 2011