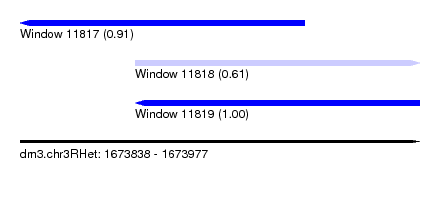
| Sequence ID | dm3.chr3RHet |
|---|---|
| Location | 1,673,838 – 1,673,977 |
| Length | 139 |
| Max. P | 0.999677 |

| Location | 1,673,838 – 1,673,937 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 63.66 |
| Shannon entropy | 0.70850 |
| G+C content | 0.56953 |
| Mean single sequence MFE | -40.10 |
| Consensus MFE | -14.18 |
| Energy contribution | -14.52 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.64 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.906734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
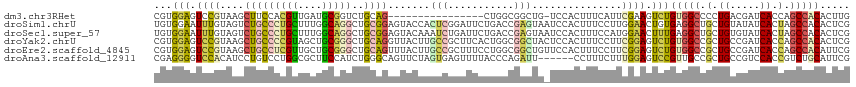
>dm3.chr3RHet 1673838 99 - 2517507 CGUGGAGUCCGUAAGCUUCCACGUUGAUGCGGUCUGCAG----------------CUGGCGGCUG-UCCACUUUCAUUCGAAGUCUGUGGCCCCUGACGAUCACCAGCCACACUUG .(((((..((((.((((.(..(((....)))....).))----------------)).))))...-))))).........((((..(((((...((.....))...))))))))). ( -29.10, z-score = 0.43, R) >droSim1.chrU 2778962 116 - 15797150 UGUGGAAUUUGUAGUCUGCCCUGCUUUGGCAGGCUGCGGAGUACCACUCGGAUUCUGACCGAGUAAUCCACUUUCCUUGGAACUGUGAGGCUGCUGUAUAUCACUAGCCACACUCG .(((((.(((((((((((((.......))))))))))))).....((((((.......))))))..))))).(((....)))..(((.(((((.((.....)).))))).)))... ( -46.50, z-score = -4.03, R) >droSec1.super_57 142527 116 + 261249 UGUGGAAUUUGUAGUCUGCCCUGCUUUGGCAGGCUGCGGAGUACAAAUCUGAUUCUGACCGAGUAAUCCACUUUCCAUGGAACUUUGAGGCUGCUGUGUAUCACUAGCCACACUCG ...(((.((((((.((((((((((....)))))..))))).))))))))).........(((((..((((.......)))).......(((((.((.....)).)))))..))))) ( -39.50, z-score = -2.46, R) >droYak2.chrU 7176401 116 + 28119190 CGUGGAGUCCGUAAGCUGCCCCGUAGCUGCGGGCUGCAGGUUACUUGCCGCUUCACUGGCGGCUACUCCACUUUCCUUCGGAGUCUGUGGCCGCUGCCGAUCACCAGCCACACUCG .(((((((..((((.(((((((((....)))))..)))).))))..((((((.....)))))).))))))).........((((..(((((.(.((.....)).).))))))))). ( -51.70, z-score = -2.88, R) >droEre2.scaffold_4845 974650 116 - 22589142 CGUGGAGUCCGUAAGCUGCCUCGUUGCUGCGGGCUGCAGUUUACUUGCCGCUUUCCUGGCGGCUGUUCCACUUUCCUUCGGAGUCUGUGGCCGCUGCCGAUCACCAGCCACAUUCG .((((((...((((((((((((((....)))))..)))))))))..((((((.....))))))..))))))....(....).(..((((((.(.((.....)).).))))))..). ( -46.60, z-score = -2.26, R) >droAna3.scaffold_12911 4274527 110 + 5364042 CGAGGGGUCCACAUCCUGUCCUGGCGCUUCCAUCUGGGCAGUUCUAGUGAGUUUUACCCAGAUU------CCUUUCUUUGGAGUCCGUUGCCGCUGCCGUCCACCGUCUGCAUUCG .((.(((..(.....(((((((((.....)))...))))))...(((((.((...((...((((------((.......)))))).)).)))))))..)..).)).))........ ( -27.20, z-score = 0.97, R) >consensus CGUGGAGUCCGUAAGCUGCCCCGUUGCUGCGGGCUGCAGAGUACUAGUCGGUUUCCUGCCGACUA_UCCACUUUCCUUCGAAGUCUGUGGCCGCUGCCGAUCACCAGCCACACUCG ...(((.((((....(((((((((....)))))..)))).......(((.(.......).)))...............)))).)))(((((.(.((.....)).).)))))..... (-14.18 = -14.52 + 0.34)

| Location | 1,673,878 – 1,673,977 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 63.11 |
| Shannon entropy | 0.59393 |
| G+C content | 0.54948 |
| Mean single sequence MFE | -37.62 |
| Consensus MFE | -14.05 |
| Energy contribution | -17.43 |
| Covariance contribution | 3.37 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.606702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3RHet 1673878 99 + 2517507 -------------------UGAAAGUGGACAGCCGCCAGCUGCAGACCGCAUCAACGUGGAAGCUUACGGACUCCACGAACGUGCAAGAGGCGGCCCUAAUGGACAGGAUCGCACACU -------------------....((((..(.((((((...((((((.....))..((((((..(.....)..))))))....))))...))))))(((.......)))...)..)))) ( -31.60, z-score = -0.90, R) >droSim1.chrU 2779002 116 + 15797150 GGAAAGUGGAUUACUCGGUCAGAAUCCGAGUGGUACU--CCGCAGCCUGCCAAAGCAGGGCAGACUACAAAUUCCACAACUGUAGAGGUGGCGAACCUACUGGAAAGAAUCGCAACCU ((((.(((((((((((((.......))))))))...)--))))((.(((((.......))))).)).....))))..........((((.((((.....((....))..)))).)))) ( -39.30, z-score = -2.32, R) >droSec1.super_57 142567 116 - 261249 GGAAAGUGGAUUACUCGGUCAGAAUCAGAUUUGUACU--CCGCAGCCUGCCAAAGCAGGGCAGACUACAAAUUCCACAACUGUAGAGGUGGCGAACCUACUGGAAAGAAUCGCAACCU ((((.(((((.(((..((((.......)))).))).)--))))((.(((((.......))))).)).....))))..........((((.((((.....((....))..)))).)))) ( -33.20, z-score = -1.19, R) >droYak2.chrU 7176441 116 - 28119190 GGAAAGUGGAGUAGCCGCCAGUGAAGCGGCAAGUAAC--CUGCAGCCCGCAGCUACGGGGCAGCUUACGGACUCCACGACUGUACAAGUGGCGGCCCUCAUGGAAAGGAUCGCACAUC .....(((((((.(((((.......)))))..((((.--((((..((((......)))))))).))))..)))))))..........((((((..(((.......)))..))).))). ( -46.40, z-score = -1.90, R) >consensus GGAAAGUGGAUUACUCGGUCAGAAUCCGACAGGUACC__CCGCAGCCCGCAAAAACAGGGCAGACUACAAACUCCACAACUGUACAAGUGGCGAACCUAAUGGAAAGAAUCGCAAACU .....(((((...(((((.......))))).........((((..((((......)))))))).........))))).............((((.(((.......))).))))..... (-14.05 = -17.43 + 3.37)



| Location | 1,673,878 – 1,673,977 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 63.11 |
| Shannon entropy | 0.59393 |
| G+C content | 0.54948 |
| Mean single sequence MFE | -44.53 |
| Consensus MFE | -23.35 |
| Energy contribution | -27.73 |
| Covariance contribution | 4.37 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.17 |
| SVM RNA-class probability | 0.999677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3RHet 1673878 99 - 2517507 AGUGUGCGAUCCUGUCCAUUAGGGCCGCCUCUUGCACGUUCGUGGAGUCCGUAAGCUUCCACGUUGAUGCGGUCUGCAGCUGGCGGCUGUCCACUUUCA------------------- ((((..((..((((.....))))((((((..((((((((.(((((((..(....).))))))).....)))...)))))..))))))))..))))....------------------- ( -39.50, z-score = -2.16, R) >droSim1.chrU 2779002 116 - 15797150 AGGUUGCGAUUCUUUCCAGUAGGUUCGCCACCUCUACAGUUGUGGAAUUUGUAGUCUGCCCUGCUUUGGCAGGCUGCGG--AGUACCACUCGGAUUCUGACCGAGUAAUCCACUUUCC ((((.((((..((.......))..)))).))))........(((((.(((((((((((((.......))))))))))))--).....((((((.......))))))..)))))..... ( -45.70, z-score = -3.88, R) >droSec1.super_57 142567 116 + 261249 AGGUUGCGAUUCUUUCCAGUAGGUUCGCCACCUCUACAGUUGUGGAAUUUGUAGUCUGCCCUGCUUUGGCAGGCUGCGG--AGUACAAAUCUGAUUCUGACCGAGUAAUCCACUUUCC ((((.((((..((.......))..)))).))))........((((((((((((.((((((((((....)))))..))))--).)))))))...((((.....))))..)))))..... ( -41.60, z-score = -3.30, R) >droYak2.chrU 7176441 116 + 28119190 GAUGUGCGAUCCUUUCCAUGAGGGCCGCCACUUGUACAGUCGUGGAGUCCGUAAGCUGCCCCGUAGCUGCGGGCUGCAG--GUUACUUGCCGCUUCACUGGCGGCUACUCCACUUUCC ..((.(((..((((.....))))..)))))...........(((((((..((((.(((((((((....)))))..))))--.))))..((((((.....)))))).)))))))..... ( -51.30, z-score = -3.25, R) >consensus AGGGUGCGAUCCUUUCCAGUAGGGCCGCCACCUCUACAGUCGUGGAAUCCGUAAGCUGCCCCGCUGUGGCAGGCUGCAG__AGUACCAGUCGGAUUCCGACCGAGUAAUCCACUUUCC ((((.(((((((((.....))))))))).))))........(((((....((((((((((.......))))))))))...........(((((.......)))))...)))))..... (-23.35 = -27.73 + 4.37)

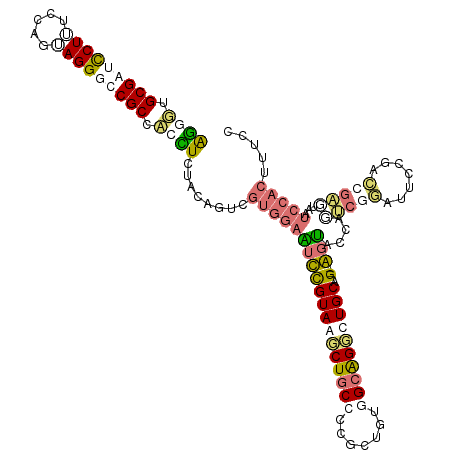

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:48:43 2011