| Sequence ID | dm3.chr3RHet |
|---|---|
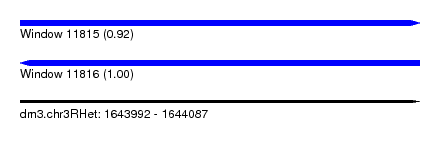
| Location | 1,643,992 – 1,644,087 |
| Length | 95 |
| Max. P | 0.997345 |

| Location | 1,643,992 – 1,644,087 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 50.87 |
| Shannon entropy | 0.70680 |
| G+C content | 0.55666 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -13.74 |
| Energy contribution | -15.63 |
| Covariance contribution | 1.90 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.921623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
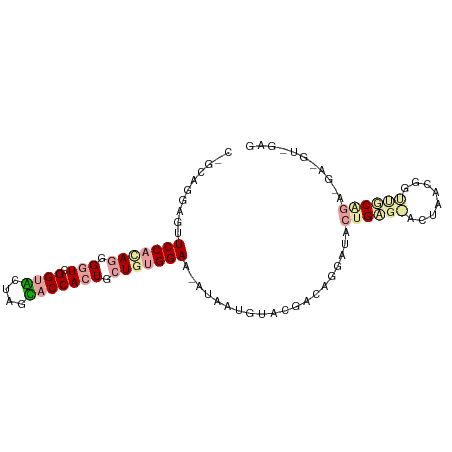
>dm3.chr3RHet 1643992 95 + 2517507 CGGCAGGUGCUCCCUAAGGAUCGGUACUUGUACCAAUCCUGUGGAACAUCAUCUACGAUGGGAUAUUGGGCAUCAACAAGCCCAUGGGA-GUAGAG .......(((((((..(((((.((((....)))).)))))(((((......))))).(((((...(((........))).)))))))))-)))... ( -35.60, z-score = -2.51, R) >droGri2.scaffold_15203 5885067 82 + 11997470 CAGCAGGAGUUCCACAGGGGUCGGUUGUAGGUCCACUGCUGUGGA---CGAGGUACU-CAGGCUUCAGCUGCCUCAGGGUUGCAGC---------- ..(((((....))...((((.((((((.((((((((....)))))---)(((...))-)...)).)))))))))).....)))...---------- ( -31.10, z-score = -0.61, R) >triCas2.chrUn_114 46873 90 - 54736 -----GGGGUUCCACAGGGGUCGGUGCUCGCACCACUGCUAUGGAAUAUAAUGUAUGACAGUGUCCUGAG-ACUGUCGGUUGCGGAAGAUGUCGAG -----...((((((.((.(((.((((....))))))).)).))))))........(((((...(((...(-((.....)))..)))...))))).. ( -30.10, z-score = -0.76, R) >consensus C_GCAGGAGUUCCACAGGGGUCGGUACUAGCACCACUGCUGUGGAA_AUAAUGUACGACAGGAUACUGAG_ACUAACGGUUGCAGA_GA_GU_GAG ..........(((((((.(((.((((....))))))).)))))))....................((((((........))))))........... (-13.74 = -15.63 + 1.90)

| Location | 1,643,992 – 1,644,087 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 50.87 |
| Shannon entropy | 0.70680 |
| G+C content | 0.55666 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -11.15 |
| Energy contribution | -12.17 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.08 |
| SVM RNA-class probability | 0.997345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3RHet 1643992 95 - 2517507 CUCUAC-UCCCAUGGGCUUGUUGAUGCCCAAUAUCCCAUCGUAGAUGAUGUUCCACAGGAUUGGUACAAGUACCGAUCCUUAGGGAGCACCUGCCG .(((((-....(((((..(((((.....))))).))))).)))))...((((((..((((((((((....))))))))))...))))))....... ( -36.80, z-score = -3.99, R) >droGri2.scaffold_15203 5885067 82 - 11997470 ----------GCUGCAACCCUGAGGCAGCUGAAGCCUG-AGUACCUCG---UCCACAGCAGUGGACCUACAACCGACCCCUGUGGAACUCCUGCUG ----------(((((.........)))))...(((..(-(((.....(---(((((....))))))(((((.........))))).))))..))). ( -26.50, z-score = -1.77, R) >triCas2.chrUn_114 46873 90 + 54736 CUCGACAUCUUCCGCAACCGACAGU-CUCAGGACACUGUCAUACAUUAUAUUCCAUAGCAGUGGUGCGAGCACCGACCCCUGUGGAACCCC----- ...................((((((-........))))))..........((((((((..((((((....)))).))..))))))))....----- ( -24.90, z-score = -1.90, R) >consensus CUC_AC_UC_CCUGCAACCGUCAGU_CCCAGAACCCUGUCGUACAUCAU_UUCCACAGCAGUGGUACAAGAACCGACCCCUGUGGAACACCUGC_G .....................(((((.......))))).............(((((((...(((((....)))))....))))))).......... (-11.15 = -12.17 + 1.01)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:48:41 2011