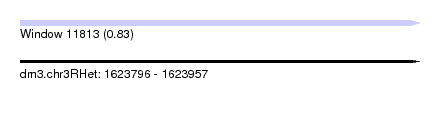
| Sequence ID | dm3.chr3RHet |
|---|---|
| Location | 1,623,796 – 1,623,957 |
| Length | 161 |
| Max. P | 0.830423 |

| Location | 1,623,796 – 1,623,957 |
|---|---|
| Length | 161 |
| Sequences | 10 |
| Columns | 181 |
| Reading direction | forward |
| Mean pairwise identity | 64.24 |
| Shannon entropy | 0.74170 |
| G+C content | 0.50300 |
| Mean single sequence MFE | -50.80 |
| Consensus MFE | -24.50 |
| Energy contribution | -24.23 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.86 |
| Mean z-score | -0.70 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.830423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3RHet 1623796 161 + 2517507 -----UGACCACUGUGCUUGCAAGACGCAGCUGCGCAAUGAGUUCGGUA---------------UAUUGAGAGGUCAUUCUUAGAAUUUGACUACCACGAAGAAUAUGACUGGCGCCAAAGUACCGUUACGAGUGAGGCGACGGCGUUGUCGCGCUGUCGCCUAGUGUAGUCCAGCUAGCG -----.......(((....)))...((((((((.((........(((((---------------(.(((.(..((((.((.((...((((.......))))...)).)).)))).)))).)))))).((((....((((((((((((....))))))))))))..)))))).))))).))) ( -54.30, z-score = -0.73, R) >droSim1.chrU 10260299 159 - 15797150 -----UGACCACUGUGCUUGCAAGACGCAGCUGCGCAAUGAGUUUGGUA---------------UAUCGUGAGGACAUUCUUAGAAUUUGACUACAACGUAGAAUAUGACUGGCGCCAAAGUACCGUUACGAGUGAGGCGACAGCGUUGUCGCGCUGUCGCCAAGUGUAGUCCAGCUAG-- -----.(((((.(((((((((.....))))..))))).)).)))((((.---------------..((.(((((....)))))))....(((((((.(((((......(((........)))....))))).....(((((((((((....)))))))))))...)))))))..)))).-- ( -52.30, z-score = -0.83, R) >droSec1.super_60 123145 159 + 163194 -----UGACCACUGUGCUUGCAAGACGCAGCUGCGCAAUGAGUUUGGUA---------------UAUCGUGAGGACAUUCUUAGAAUUUGACUACAACGUAGAAUAUGACUGGCGCCAAAGUACCGUUACGAGUGAGGCGACGGCGUUGUCGCGCUGUCGCCAAGUGUAGUCCAGCUAG-- -----.(((((.(((((((((.....))))..))))).)).)))((((.---------------..((.(((((....)))))))....(((((((.(((((......(((........)))....))))).....(((((((((((....)))))))))))...)))))))..)))).-- ( -51.60, z-score = -0.39, R) >droYak2.chr3L 442970 159 - 24197627 -----UGACCACUGCGCGUGCAAGACGCAGCUGCGCAAUAAGUUUGGU---------------UUAUUGAGAACUUCUUCU--AUAUUUGACUACGACGCAUAAUAUGACUGGCGCCAAAGUACCGAAAUGAGUGAGGCGACGGCGUUGUCGCGCUGUCGCCAAGUGUAGUCCAGCUAGCG -----........((((((.....))))(((((..(((((((.....)---------------))))))((((....))))--......(((((((((.(((......(((........)))......))).))..(((((((((((....)))))))))))...)))))))))))).)). ( -50.40, z-score = -0.06, R) >droEre2.scaffold_4929 21595141 176 + 26641161 -----UGACCACUGCGCUUGCAAGACGCUGUUGCGCAAUGAGUUUGGAAAUUGUGAAGUUGAGUUAUUGUGAAAUCAUUCUUAGAAUUUGACUACUACGCAGAAUAUGACUGGCGCCAAAGUACCGAAACGAGUGAGGCGACGGCGUUGUCGCGCUGUCGACAAGUGUAGUCCAGCUAGCG -----.....(((((((((....(((((((((((.((.....(((((...(((((.(((..((((...(((....)))......))))..)))....)))))......(((........))).))))).....))..)))))))))))((((......))))))))))))).......... ( -49.90, z-score = 0.75, R) >droAna3.scaffold_13339 3523753 157 + 4599533 UCACCAGAGUGAUAAUAUUGCAUCAACAUCUUGCGCUGUGAGUGUGUGG-----------------UUGCCAGGCAACCGUUGGCAGGCGACCGUUGCUUGGAUGUUGUCGCUUUACAAAGUACCGUUGU-AGCGAGGCGACAGCGUUGUGUUGUUGUCGCCGAUCGAGGUUUGG------ ..((((((((((((((((((((...((((((..(...)..)).))))..-----------------.)))((((((((.((((.....)))).)))))))))))))))))))))).........(((...-.))).((((((((((......))))))))))......)))....------ ( -60.10, z-score = -2.01, R) >droWil1.scaffold_181143 435578 155 + 1323777 -----UGACCACUGUGCUUGCCAGAGCAAGCUGUGCAAUGAGUUUGGGA---------------UAUUGGGAAUGC-UUCUUAGAAUUUGACUAUAGCGAAGAAUAUGACUGGCGCCUAGGUGCCGU-ACGAGUGAGGCGACAGCGUUGUCGCGCUGUUGCCGAGUGUAGUCCAGCU---- -----.(((((((..((((((....))))))(((((....((((..(((---------------(.((((((....-.)))))).))))))))..................(((((....)))))))-))))))).(((((((((((....))))))))))).......))).....---- ( -54.00, z-score = -1.23, R) >droVir3.scaffold_13324 1920124 160 - 2960039 -----UGACCACUGUGCUUGCAAGACGCUGCUGCGCAAUGUGUUUGGCA---------------AUUUGUGAAAUCAUUCUUAGAAUUUGACUAUAACGUAGAAUAUGACUGGCGCCAAAAUACCGU-ACAAGUGAGGGGACGGCGUUGUCGCGCUGUCCCCGAGUGUAGUCCAGCUGGCG -----.(.((((((..((.(((...((((..((((....((((((((((---------------((((..(((....)))...)))))...((((...))))............))).)))))))))-)..)))).(((((((((((....)))))))))))...)))))..))).)))). ( -51.10, z-score = -0.78, R) >droMoj3.scaffold_6500 31235632 142 + 32352404 ------------UGUGCUUGUAUGUCGCUGCUGUGCAAUAGAUUUGGUC---------------UAUUAUGAAAU--UUUUUAGAAAUUC-CAAGAAUUAUGAAUAUCGGUGGCGCCC---UUUC------AGCGAGGUGACGGCGGUGUUGCGCUGUCGCUGAGUGUAGUCCAACUUGCG ------------((..(((.....((((((..(((((((((((...)))---------------)))).......--......((.((((-..........)))).))....))))..---...)------)))))((((((((((......)))))))))))))..))............ ( -38.80, z-score = 0.08, R) >droGri2.scaffold_15074 6719306 140 + 7742996 --------------UAACCCAGACCACCUACCCCGCCGCCAGGCAGCUGU-------------UUGCUGUACUUCAAGUGUCCCAGUAUUAGCACAAAGGAUAAUGCUGCUGUUAC--AAUUGCCAUUGCUGGCA--GUUGUAGCGAU-UGUUGCGAGUGCUGUUUCUGACG--------- --------------.....((((...........(((....)))(((...-------------..)))(((((((((..(((.((((((((.(......).))))))))..(((((--((((((((....)))))--)))))))))))-..))).))))))....))))...--------- ( -45.50, z-score = -1.85, R) >consensus _____UGACCACUGUGCUUGCAAGACGCAGCUGCGCAAUGAGUUUGGUA_______________UAUUGUGAAGUCAUUCUUAGAAUUUGACUACAACGUAGAAUAUGACUGGCGCCAAAGUACCGUUACGAGUGAGGCGACGGCGUUGUCGCGCUGUCGCCGAGUGUAGUCCAGCUAG__ ............(((((..((........)).)))))....................................................((((((.((.......................(((........))).((((((((((......))))))))))..))))))))......... (-24.50 = -24.23 + -0.27)

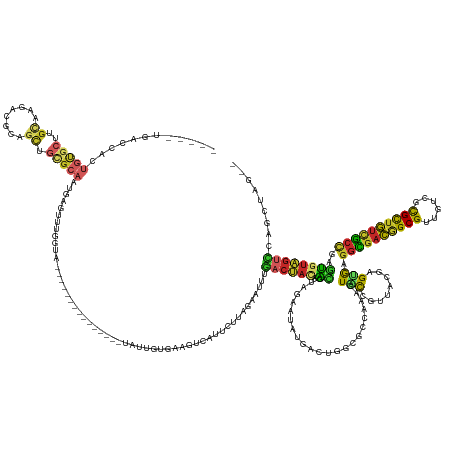
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:48:39 2011