| Sequence ID | dm3.chr3RHet |
|---|---|
| Location | 1,558,844 – 1,558,935 |
| Length | 91 |
| Max. P | 0.877451 |

| Location | 1,558,844 – 1,558,935 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 74.15 |
| Shannon entropy | 0.51993 |
| G+C content | 0.45817 |
| Mean single sequence MFE | -27.74 |
| Consensus MFE | -15.38 |
| Energy contribution | -15.90 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.877451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
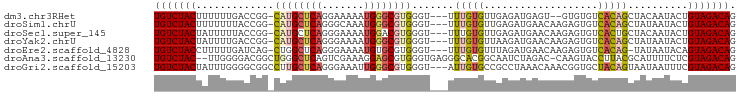
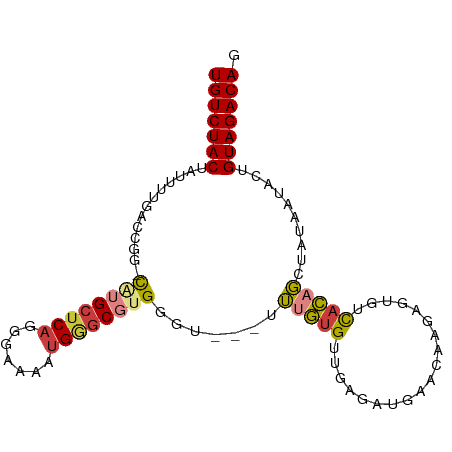
>dm3.chr3RHet 1558844 91 - 2517507 UGUCUACUUUUUUGACCGG-CAUGCUCAGGAAAAAUGGGCGUGGGU---UUUGUGUUGAGAUGAGU--GUGUGUCACAGCUACAAUACUGUAGACAG (((((((............-((((((((.......))))))))(((---.((((((((.(((....--....))).)))).)))).)))))))))). ( -30.50, z-score = -2.46, R) >droSim1.chrU 9946624 93 + 15797150 UGUCUACUUUUUUUACCGG-CAUGCUCAGGGCAAAUGGGCGUGGGU---UUUGUGUUGAGAUGAACAAGAGUGUCACAGCUAUAAUACUGUAGACAG (((((((............-((((((((.......))))))))(((---.((((((((.((((........)))).)))).)))).)))))))))). ( -27.90, z-score = -1.74, R) >droSec1.super_145 21386 93 - 49412 UGUCUACUAUUUUUACCGG-CAUGCUCAGGGAAAAUGGACGUGGGU---UUUGUGUUGAGAUGAACAAGAGUGUCACUGCUACAAUACUGUAGACAG (((((((.......(((..-((((..((.......))..)))))))---...((((((((.(((((....)).)))...)).)))))).))))))). ( -22.30, z-score = -0.19, R) >droYak2.chrU 12659620 93 - 28119190 UGUCUACUAUUUUGACCGG-CAUGCUCAGGGAAAAUGGGCGUGGGU---UUUGUGUUAAGAUGAACAAGAGUGUCACAGCUAUAAUACUGUAGACAG (((((((......((((..-((((((((.......)))))))))))---)..((((((((.(((((....)).)))...)).)))))).))))))). ( -26.80, z-score = -1.87, R) >droEre2.scaffold_4828 85148 92 - 157145 UGUCUACCUUUUUGAUCAG-CUGGCUCAGGGAAAAUGUGCGUGGGU---UUUGUGUUUAGAUGAACAAGAGUGUCACAG-UAUAAUACAGUAGACAG (((((((.(((((((((..-..)).)))))))...(((((((((..---(((.(((((....))))).)))..)))).)-)))).....))))))). ( -22.60, z-score = -0.56, R) >droAna3.scaffold_13230 335900 94 + 3602488 UGUCUAC--UUGGGGACGGCUGGGCUCAGUCGAAAGGAGCGUGGGUGAGGGCACGGCAAUCUAGAC-CAAGUACCUUACGCAUUUUCUCGUAGACAG (((((((--.......((((((....))))))...((((.(((.((((((((..(((......).)-)..)).)))))).))).)))).))))))). ( -33.30, z-score = -1.69, R) >droGri2.scaffold_15203 1884788 94 - 11997470 UGUCUACUAUUUGGGGCGGCCUUGCUCAGGGAAAUUGGGCGUGGGU---AUUGUGCCGCCUAAACAAACGGUGCUACAGUAAUAAUUUCGUAGACAG (((((((......((((((((.((((((.(.........).)))))---)..).)))))))..........(((....)))........))))))). ( -30.80, z-score = -1.71, R) >consensus UGUCUACUAUUUUGACCGG_CAUGCUCAGGGAAAAUGGGCGUGGGU___UUUGUGUUGAGAUGAACAAGAGUGUCACAGCUAUAAUACUGUAGACAG (((((((.............((((((((.......)))))))).......(((((...................)))))..........))))))). (-15.38 = -15.90 + 0.51)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:48:36 2011