| Sequence ID | dm3.chr3RHet |
|---|---|
| Location | 1,429,610 – 1,429,670 |
| Length | 60 |
| Max. P | 0.999408 |

| Location | 1,429,610 – 1,429,670 |
|---|---|
| Length | 60 |
| Sequences | 4 |
| Columns | 60 |
| Reading direction | forward |
| Mean pairwise identity | 64.04 |
| Shannon entropy | 0.59747 |
| G+C content | 0.45065 |
| Mean single sequence MFE | -17.43 |
| Consensus MFE | -12.14 |
| Energy contribution | -13.70 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.33 |
| SVM RNA-class probability | 0.998339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
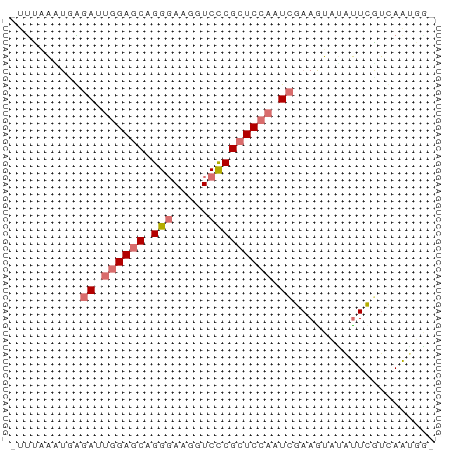
>dm3.chr3RHet 1429610 60 + 2517507 UUUAGGGAGAGAUUUGAGCAGGCAAGCUCCCGCUCUAAUCGAAGUAUCUUUUGUAAGUGU ...(((((..((((.((((.((......)).)))).))))......)))))......... ( -11.80, z-score = 0.43, R) >droSim1.chrU 13726799 56 - 15797150 -UUUAAUUUGGA--GGAGCAGGGAAGGUCCCGCUCCAUUCGAAGUUAAUUCGUCAAUGU- -.(((((((.((--(((((.(((.....))))))))..)).)))))))...........- ( -19.30, z-score = -3.27, R) >droSec1.super_61 95801 58 - 189864 -UUUAAAUGAGAUUGGAUCAGGGAAGGUCCCGCUCCAAUCGAAGUAUACUCGUGAGAGA- -.........(((((((...(((.....)))..)))))))........(((....))).- ( -21.00, z-score = -3.34, R) >droEre2.scaffold_4929 4411843 59 - 26641161 -ACCUGCUCGUAGUGGAGCAGGGAAGGUCUCGCUCCAGUUGAAAUAUAUUCGUCAAUUGU -.(((((((......))))))).............(((((((..........))))))). ( -17.60, z-score = -1.99, R) >consensus _UUUAAAUGAGAUUGGAGCAGGGAAGGUCCCGCUCCAAUCGAAGUAUAUUCGUCAAUGG_ ..........(((((((((.(((.....)))))))))))).................... (-12.14 = -13.70 + 1.56)

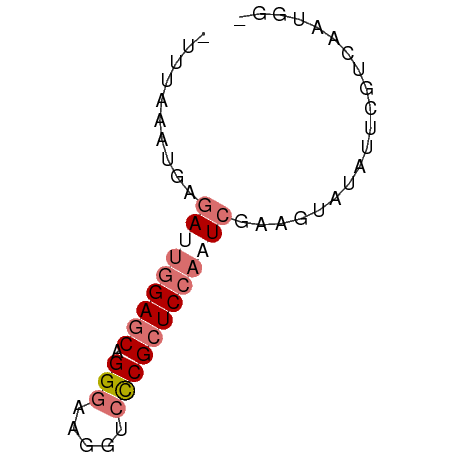
| Location | 1,429,610 – 1,429,670 |
|---|---|
| Length | 60 |
| Sequences | 4 |
| Columns | 60 |
| Reading direction | reverse |
| Mean pairwise identity | 64.04 |
| Shannon entropy | 0.59747 |
| G+C content | 0.45065 |
| Mean single sequence MFE | -15.80 |
| Consensus MFE | -13.50 |
| Energy contribution | -15.25 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.86 |
| SVM RNA-class probability | 0.999408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3RHet 1429610 60 - 2517507 ACACUUACAAAAGAUACUUCGAUUAGAGCGGGAGCUUGCCUGCUCAAAUCUCUCCCUAAA ....................((((.(((((((......))))))).)))).......... ( -15.40, z-score = -2.71, R) >droSim1.chrU 13726799 56 + 15797150 -ACAUUGACGAAUUAACUUCGAAUGGAGCGGGACCUUCCCUGCUCC--UCCAAAUUAAA- -...(((.((((.....))))...((((((((......))))))))--..)))......- ( -17.70, z-score = -4.30, R) >droSec1.super_61 95801 58 + 189864 -UCUCUCACGAGUAUACUUCGAUUGGAGCGGGACCUUCCCUGAUCCAAUCUCAUUUAAA- -.(((....)))........(((((((.((((......)))).))))))).........- ( -16.50, z-score = -2.60, R) >droEre2.scaffold_4929 4411843 59 + 26641161 ACAAUUGACGAAUAUAUUUCAACUGGAGCGAGACCUUCCCUGCUCCACUACGAGCAGGU- .........(((.....)))....((((......))))(((((((......))))))).- ( -13.60, z-score = -1.95, R) >consensus _CAAUUGACGAAUAUACUUCGAUUGGAGCGGGACCUUCCCUGCUCCAAUCCCAACUAAA_ ....................((((((((((((......)))))))))))).......... (-13.50 = -15.25 + 1.75)
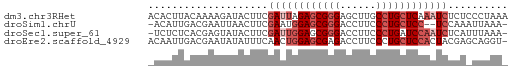
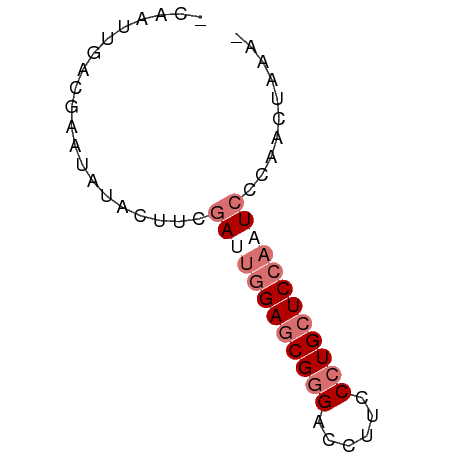
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:48:32 2011