
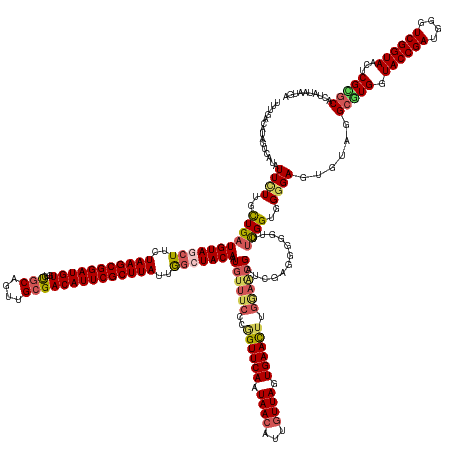
| Sequence ID | dm3.chr3RHet |
|---|---|
| Location | 1,412,292 – 1,412,479 |
| Length | 187 |
| Max. P | 0.997337 |

| Location | 1,412,292 – 1,412,479 |
|---|---|
| Length | 187 |
| Sequences | 5 |
| Columns | 187 |
| Reading direction | forward |
| Mean pairwise identity | 87.10 |
| Shannon entropy | 0.23793 |
| G+C content | 0.46407 |
| Mean single sequence MFE | -63.34 |
| Consensus MFE | -50.66 |
| Energy contribution | -52.14 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.983702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3RHet 1412292 187 + 2517507 UUUGACAUAGUCAUAUUCUUGUUGUUGUAGCUUCUAAGCGGAUGUGUCUGCAGUUGCGACAUUCGCUUAUUGGCUACAAUGUUUCCCGGUUCAAUAAAAUUCUUAGUGAACGUGAAACGAUCGAGGGGGUUUCGGUGGGGAGUGUAGGCGUGUUACCGAUGGGUCGGUAACUCGCGCACUAUAAUGA ......((((((((((((((...(((((((((..(((((((((((...(((....))))))))))))))..)))))))))(((((.((.((((.(((.....))).)))))).))))).((((((.....)))))).))))))))..(((.((((((((....)))))))).)))).)))))..... ( -68.10, z-score = -3.85, R) >droEre2.scaffold_4845 22547886 183 - 22589142 -UUGACAUAGU---AUUCUUGCUGAUGUAGCUUCUAAGCGGAUGUGUCUGCAGUUGCGACAUUCGCUUAUUAGCUACAAUGUUUCCCGGUUCAAUAACAUUGUUAGUGAAUUUGGAACGAUCGAGGGGGUUUCGGUGGAGAGUGUAGGCGUGGUACCGAUGGGUCGGUAACUCGUGCGUUGUUGUGA -....((((((---(((((..(((((((((((..(((((((((((...(((....))))))))))))))..))))))).((((.((..(((((.((((...)))).)))))..))))))............))))..).)))))...(((.(.((((((....)))))).).)))......))))). ( -61.00, z-score = -2.01, R) >droYak2.chr2L_random 2740495 187 - 4064425 UUUGACAUAGUCAUAUUUUUGCUGAUGUAGCUUCUAAGCGGAUGUGUCUGCAGUUGCGACAUUCGCUUAUUAACUACAAUGUUUCCCGGUUCAAUAACAUUGUUAGUGAACUUGGAACGAUCGAGGGGGUUUCGGUGGGGAGUGUAGGCAUGGUACCGAUGGGUCGGUAACUCGUGCGAAAUGUGGA ..........(((((((((......(((((.(..(((((((((((...(((....))))))))))))))..).)))))(..((((((((((((.((((...)))).))))))((((((..(....)..))))))..))))))..)..((((((((((((....))))).)).)))))))))))))). ( -60.50, z-score = -1.94, R) >droSec1.super_81 62816 178 + 123496 UUUGACCUAGUCAUAUUCUUGCUGAUGUAGCUUCUAAGCGGAUGUGUCUGCAGUUGAGACAUUCGCUUAUUGGAUACAAUGUUUCCCGGUUCAAUAACAUUGUUAGUGAACUUGGAGCGAUCGUGGGGGUUUUGGUGGGGAGUGUAGGCGUGGUACCGAUGGGUCGGUAACGCGACCA--------- .....(((..(((.(((((..(.((((((.((..(((((((((((.((.......)).)))))))))))..)).)))..((((.((.((((((.((((...)))).)))))).))))))))))..)))))..)))..)))......((((((.((((((....)))))).)))).)).--------- ( -65.30, z-score = -3.09, R) >droSim1.chrU 6331469 179 - 15797150 UUUGACAUAAUCAUAUUCUUGCUGAUGUAGCUUCUAAGCGGAUGUGUCCGCAGUUGCGACAUUCGCUUAUUGGAUACAUUA------AGUUCAAUAACAUUGUUAGUGAACUUGGAGCGAUCGUGGGGGUUUCGGUGGGGAGUGUAUGCGUGGUACCGAUGGGUCGGUAACUCGCGCACCCGAAU-- ..........((...((((..((((...(((((((..(((((....)))))....((((...((((((..((....)).((------((((((.((((...)))).)))))))))))))))))))))))))))))..))))..(..(((((((((((((....))))).)).))))))..)))..-- ( -61.80, z-score = -2.23, R) >consensus UUUGACAUAGUCAUAUUCUUGCUGAUGUAGCUUCUAAGCGGAUGUGUCUGCAGUUGCGACAUUCGCUUAUUGGCUACAAUGUUUCCCGGUUCAAUAACAUUGUUAGUGAACUUGGAACGAUCGAGGGGGUUUCGGUGGGGAGUGUAGGCGUGGUACCGAUGGGUCGGUAACUCGCGCACUAUAAUGA ...............((((..(((((((((((..(((((((((((...(((....))))))))))))))..)))))))....((((.((((((.((((...)))).)))))).))))..............))))..))))......(((((.((((((....))))))...))))).......... (-50.66 = -52.14 + 1.48)

| Location | 1,412,292 – 1,412,479 |
|---|---|
| Length | 187 |
| Sequences | 5 |
| Columns | 187 |
| Reading direction | reverse |
| Mean pairwise identity | 87.10 |
| Shannon entropy | 0.23793 |
| G+C content | 0.46407 |
| Mean single sequence MFE | -47.04 |
| Consensus MFE | -31.72 |
| Energy contribution | -31.96 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.81 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.08 |
| SVM RNA-class probability | 0.997337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3RHet 1412292 187 - 2517507 UCAUUAUAGUGCGCGAGUUACCGACCCAUCGGUAACACGCCUACACUCCCCACCGAAACCCCCUCGAUCGUUUCACGUUCACUAAGAAUUUUAUUGAACCGGGAAACAUUGUAGCCAAUAAGCGAAUGUCGCAACUGCAGACACAUCCGCUUAGAAGCUACAACAACAAGAAUAUGACUAUGUCAAA ........(((.(((.((((((((....)))))))).))).)))..((((....(((((..........)))))..(((((.((((...)))).))))).))))....(((((((...((((((..((((((....)).))))....))))))...)))))))...........((((...)))).. ( -54.50, z-score = -5.71, R) >droEre2.scaffold_4845 22547886 183 + 22589142 UCACAACAACGCACGAGUUACCGACCCAUCGGUACCACGCCUACACUCUCCACCGAAACCCCCUCGAUCGUUCCAAAUUCACUAACAAUGUUAUUGAACCGGGAAACAUUGUAGCUAAUAAGCGAAUGUCGCAACUGCAGACACAUCCGCUUAGAAGCUACAUCAGCAAGAAU---ACUAUGUCAA- ..........((.((((.((((((....))))))............((......))......))))....((((...((((.((((...)))).))))...))))....(((((((..((((((..((((((....)).))))....))))))..)))))))...))......---..........- ( -43.40, z-score = -3.54, R) >droYak2.chr2L_random 2740495 187 + 4064425 UCCACAUUUCGCACGAGUUACCGACCCAUCGGUACCAUGCCUACACUCCCCACCGAAACCCCCUCGAUCGUUCCAAGUUCACUAACAAUGUUAUUGAACCGGGAAACAUUGUAGUUAAUAAGCGAAUGUCGCAACUGCAGACACAUCCGCUUAGAAGCUACAUCAGCAAAAAUAUGACUAUGUCAAA ..........(((((((.((((((....)))))).).................(((.......))).))))(((..(((((.((((...)))).)))))..))).....(((((((..((((((..((((((....)).))))....))))))..)))))))...)).......((((...)))).. ( -46.00, z-score = -3.76, R) >droSec1.super_81 62816 178 - 123496 ---------UGGUCGCGUUACCGACCCAUCGGUACCACGCCUACACUCCCCACCAAAACCCCCACGAUCGCUCCAAGUUCACUAACAAUGUUAUUGAACCGGGAAACAUUGUAUCCAAUAAGCGAAUGUCUCAACUGCAGACACAUCCGCUUAGAAGCUACAUCAGCAAGAAUAUGACUAGGUCAAA ---------(((((((((((((((....))))))..)))).............................(((((..(((((.((((...)))).)))))..))).....((((.(...((((((.((((.((.......)).)))).))))))...).))))...))........)))))....... ( -43.10, z-score = -2.68, R) >droSim1.chrU 6331469 179 + 15797150 --AUUCGGGUGCGCGAGUUACCGACCCAUCGGUACCACGCAUACACUCCCCACCGAAACCCCCACGAUCGCUCCAAGUUCACUAACAAUGUUAUUGAACU------UAAUGUAUCCAAUAAGCGAAUGUCGCAACUGCGGACACAUCCGCUUAGAAGCUACAUCAGCAAGAAUAUGAUUAUGUCAAA --(((((((((((((.(.((((((....)))))).).)))..................................(((((((.((((...)))).))))))------)...)))))).....((..((((.((..(((((((....)))))..))..)).))))..))..)))).((((...)))).. ( -48.20, z-score = -3.35, R) >consensus UCAUUAUAGUGCACGAGUUACCGACCCAUCGGUACCACGCCUACACUCCCCACCGAAACCCCCUCGAUCGUUCCAAGUUCACUAACAAUGUUAUUGAACCGGGAAACAUUGUAGCCAAUAAGCGAAUGUCGCAACUGCAGACACAUCCGCUUAGAAGCUACAUCAGCAAGAAUAUGACUAUGUCAAA .............((.(.((((((....)))))).).))..............................(((((..(((((.((((...)))).)))))..))).....((((((...((((((..((((((....)).))))....))))))...))))))...)).................... (-31.72 = -31.96 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:48:30 2011