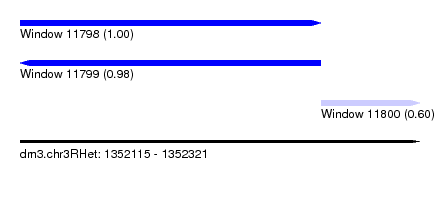
| Sequence ID | dm3.chr3RHet |
|---|---|
| Location | 1,352,115 – 1,352,321 |
| Length | 206 |
| Max. P | 0.997354 |

| Location | 1,352,115 – 1,352,270 |
|---|---|
| Length | 155 |
| Sequences | 5 |
| Columns | 174 |
| Reading direction | forward |
| Mean pairwise identity | 84.27 |
| Shannon entropy | 0.28519 |
| G+C content | 0.47954 |
| Mean single sequence MFE | -52.08 |
| Consensus MFE | -37.81 |
| Energy contribution | -38.97 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.997354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
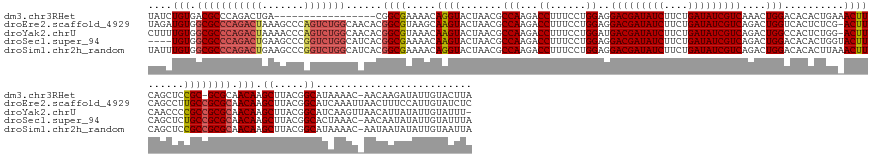
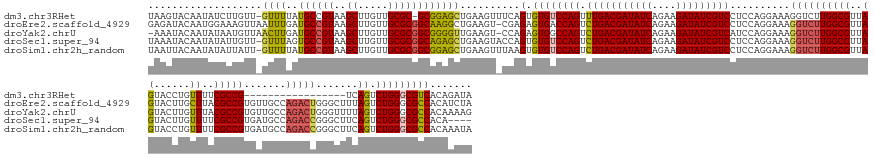
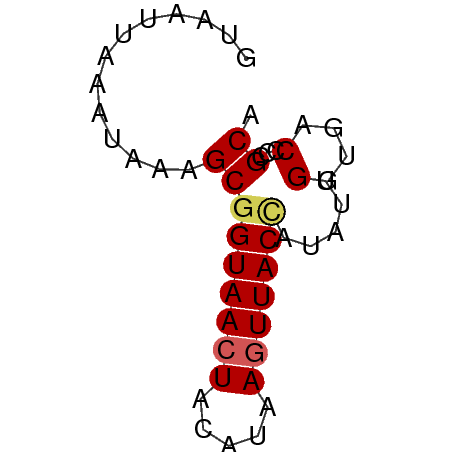
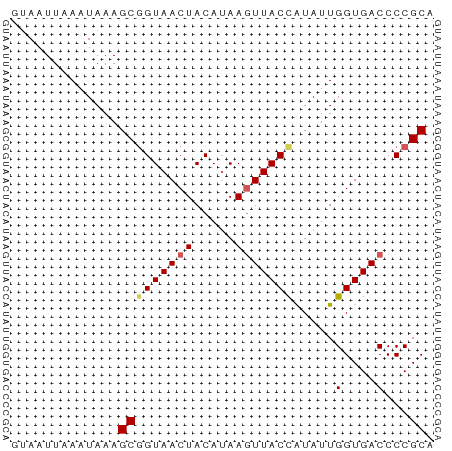
>dm3.chr3RHet 1352115 155 + 2517507 UAUCUGUGACGCCCAGACUGA-----------------CGGCGAAAACAGGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAAACUGGACACACUGAAACUUCAGCUCCGC-GCGCAACAAGCUUACGGCAUAAAAC-AACAAGAUAUUGUACUUA (((((((..((((........-----------------.))))...))))))).....(((............(((((((((((((....)))))))))............((((....))))))))..-((.......))....))).......-.((((....))))..... ( -42.80, z-score = -3.17, R) >droEre2.scaffold_4929 4656532 173 - 26641161 UAGAUGUGGCGCCCAGACUAAAGCCCAGUCUGGCAACACGGCGUAAGCAAGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGUCACUCUCG-ACUUCAGCCUUGCCGCGCAACAAGCUUACGGCAUCAAAUUAACUUUCCAUUGUAUCUC .(((((.((((((.(((((.......)))))))).....(((((.((......)).)))))..((((..((......(((((((((....))))))))).))..)))).......-......))).(((((.((.....))...)))))...................))))). ( -54.00, z-score = -2.72, R) >droYak2.chrU 2414590 172 + 28119190 CUUUUGUGGCGCCCAGACUAAAACCCAGUCUGGCAACACGGCGUAAACAAGUACUAACGCCAAGACCUUUCCUGGAUGACGAUAUCUUCUGAUAUCGUCAGACUGGCCACUCUGG-ACUUCAACCCCGCCGCGCAACAAGCUUACGGCAUCAAGUUAACAUUAUAUUGUAUUU- ...((((.(((((((((((.......)))))))......((((.....((((.(((..((((...((......)).((((((((((....))))))))))...)))).....)))-))))......)))))))).))))((.....)).........(((......)))....- ( -51.90, z-score = -3.59, R) >droSec1.super_94 45069 169 - 93855 ----UGUGGCGCCCAGACUGAAGCCCGGUCUGGCAUCACGGCGAAAACAAGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGACACACUGGUACUUCAGCUCUGCCGCGCAACAAGCUUACGGCACUAAAC-AACAAUAUAUUGUAUUUA ----(((.((((.((((((((((.((((((((((......((........))......))).)))))..(((.(...(((((((((....)))))))))...).)))......))..)))))).))))..)))).))).((.....)).......-.((((....))))..... ( -55.30, z-score = -3.32, R) >droSim1.chr2h_random 683813 173 + 3178526 UAUUUGUGGCGCCCAGACUGAAGCCCGGUCUGGCAUCACGGCGAAAACAGGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGACACACUUAAACUUCAGCUCCGCCGCGCAACAAGCUUACGGCAUAAAAC-AAUAAUAUAUUGUAAUUA ...((((.((((((((((((.....))))))))......((((((...((((.((.......)))))))))..(((((((((((((....)))))))))...(((((...........)))))))))))))))).))))((.....)).....((-((((...))))))..... ( -56.40, z-score = -4.16, R) >consensus UAUUUGUGGCGCCCAGACUGAAGCCCAGUCUGGCAACACGGCGAAAACAAGUACUAACGCCAAGACCUUUCCUGGAGGACGAUAUCUUCUGAUAUCGUCAGACUGGACACACUGG_ACUUCAGCUCCGCCGCGCAACAAGCUUACGGCAUAAAAC_AACAAUAUAUUGUAUUUA ....(((.(((((((((((.......)))))))......((((.....((((.......(((...((......))..(((((((((....)))))))))....)))..........))))......)))))))).))).((.....)).......................... (-37.81 = -38.97 + 1.16)
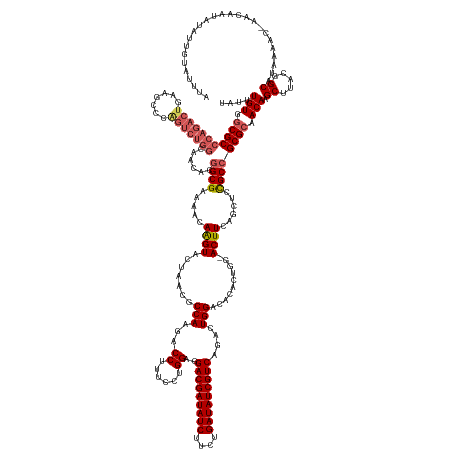
| Location | 1,352,115 – 1,352,270 |
|---|---|
| Length | 155 |
| Sequences | 5 |
| Columns | 174 |
| Reading direction | reverse |
| Mean pairwise identity | 84.27 |
| Shannon entropy | 0.28519 |
| G+C content | 0.47954 |
| Mean single sequence MFE | -58.72 |
| Consensus MFE | -41.97 |
| Energy contribution | -42.09 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.975308 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
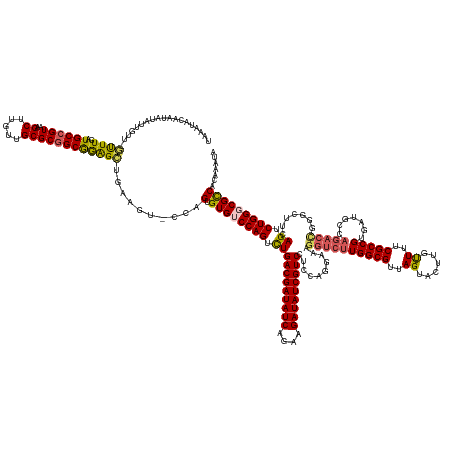
>dm3.chr3RHet 1352115 155 - 2517507 UAAGUACAAUAUCUUGUU-GUUUUAUGCCGUAAGCUUGUUGCGC-GCGGAGCUGAAGUUUCAGUGUGUCCAGUUUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACCUGUUUUCGCCG-----------------UCAGUCUGGGCGUCACAGAUA ...(..(((((.((((..-((.....))..))))..)))))..)-..(((((....))))).((((((((((.(((((((((((....)))))).(((....)))........((((..((......))..)))))-----------------)))).))))))).)))..... ( -47.70, z-score = -1.87, R) >droEre2.scaffold_4929 4656532 173 + 26641161 GAGAUACAAUGGAAAGUUAAUUUGAUGCCGUAAGCUUGUUGCGCGGCAAGGCUGAAGU-CGAGAGUGACCAGUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACUUGCUUACGCCGUGUUGCCAGACUGGGCUUUAGUCUGGGCGCCACAUCUA (((((....((((............((((((..((.....))))))))((((((..((-((....))))))))))(((((((((....))))))))).))))......)))))(((((.(((....))).)))))(((.((((((((((.....)))))).)))).)))..... ( -65.90, z-score = -3.04, R) >droYak2.chrU 2414590 172 - 28119190 -AAAUACAAUAUAAUGUUAACUUGAUGCCGUAAGCUUGUUGCGCGGCGGGGUUGAAGU-CCAGAGUGGCCAGUCUGACGAUAUCAGAAGAUAUCGUCAUCCAGGAAAGGUCUUGGCGUUAGUACUUGUUUACGCCGUGUUGCCAGACUGGGUUUUAGUCUGGGCGCCACAAAAG -...............((((((...((((((..((.....)))))))).)))))).((-(((((.(((((((((((((((((((....)))))))))..........(((..((((((............))))))....))))))))))...))).))))))).......... ( -62.30, z-score = -3.16, R) >droSec1.super_94 45069 169 + 93855 UAAAUACAAUAUAUUGUU-GUUUAGUGCCGUAAGCUUGUUGCGCGGCAGAGCUGAAGUACCAGUGUGUCCAGUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACUUGUUUUCGCCGUGAUGCCAGACCGGGCUUCAGUCUGGGCGCCACA---- (((((((((....))).)-))))).((((((..((.....))))))))..((((......))))((((((((.(((((((((((....)))))))(((....)))..(((((.(((((((................)))))))))))).....)))).))))))))....---- ( -59.79, z-score = -2.56, R) >droSim1.chr2h_random 683813 173 - 3178526 UAAUUACAAUAUAUUAUU-GUUUUAUGCCGUAAGCUUGUUGCGCGGCGGAGCUGAAGUUUAAGUGUGUCCAGUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACCUGUUUUCGCCGUGAUGCCAGACCGGGCUUCAGUCUGGGCGCCACAAAUA .....((((((...))))-))....((((((..((.....))))))))((((....))))..((((((((((.(((((((((((....)))))))(((....)))..(((((.(((((((................)))))))))))).....)))).))))))).)))..... ( -57.89, z-score = -2.33, R) >consensus UAAAUACAAUAUAUUGUU_GUUUGAUGCCGUAAGCUUGUUGCGCGGCGGAGCUGAAGU_CCAGUGUGUCCAGUCUGACGAUAUCAGAAGAUAUCGUCCUCCAGGAAAGGUCUUGGCGUUAGUACUUGUUUUCGCCGUGAUGCCAGACCGGGCUUCAGUCUGGGCGCCACAAAUA ...................((((..((((((..((.....))))))))))))..........(.((((((((.(((((((((((....)))))))))..........((((((((((..((......))..))))).......))))).......)).)))))))))....... (-41.97 = -42.09 + 0.12)
| Location | 1,352,270 – 1,352,321 |
|---|---|
| Length | 51 |
| Sequences | 5 |
| Columns | 51 |
| Reading direction | forward |
| Mean pairwise identity | 92.94 |
| Shannon entropy | 0.12301 |
| G+C content | 0.37255 |
| Mean single sequence MFE | -10.98 |
| Consensus MFE | -8.66 |
| Energy contribution | -8.90 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.595078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
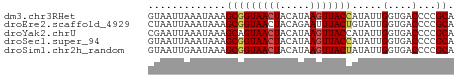
>dm3.chr3RHet 1352270 51 + 2517507 GUAAUUAAAUAAAGCGGUAACUACAUAAGUUACCAUAUUGGUGACCCCGCA .............(((((((((.....))))))).....(....)...)). ( -12.20, z-score = -2.23, R) >droEre2.scaffold_4929 4656705 51 - 26641161 CUAAUUAAAUAAAGCGGUAACUACAGAAUUUACUGUAUUGGUGACCCCGCA .............((((....(((((......)))))..(....).)))). ( -11.50, z-score = -2.16, R) >droYak2.chrU 2414762 51 + 28119190 CGAAUUAAAUAAAGCAGUAACUACAUAAGUUACCAUAUUGGUGACCCCGCA .............((.((....))....((((((.....))))))...)). ( -8.90, z-score = -1.97, R) >droSec1.super_94 45238 51 - 93855 GUAAUUAAAUAAAGCGGUAACUACAUAAGUUACCAUAUUGGUGACCCCGCA .............(((((((((.....))))))).....(....)...)). ( -12.20, z-score = -2.23, R) >droSim1.chr2h_random 683986 51 + 3178526 GUAAUUGAAUAAAGCGGUAACUACAUAAGUUACUAUAUUGGUGACCCCGCA .............(((((((((.....))))))......(....)..))). ( -10.10, z-score = -1.15, R) >consensus GUAAUUAAAUAAAGCGGUAACUACAUAAGUUACCAUAUUGGUGACCCCGCA .............(((((((((.....))))))).....((.....)))). ( -8.66 = -8.90 + 0.24)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:48:27 2011