| Sequence ID | dm3.chr3RHet |
|---|---|
| Location | 1,162,807 – 1,162,908 |
| Length | 101 |
| Max. P | 0.953950 |

| Location | 1,162,807 – 1,162,908 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 54.16 |
| Shannon entropy | 0.75989 |
| G+C content | 0.51081 |
| Mean single sequence MFE | -35.58 |
| Consensus MFE | -10.20 |
| Energy contribution | -10.00 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.75 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.907184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
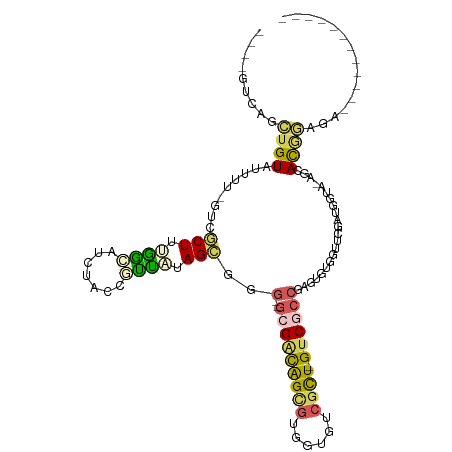
>dm3.chr3RHet 1162807 101 + 2517507 AUUGGUCGGCUGUAUUUUUGUAGCUUUGGCAUCUCCCGUUACAGCGCG--GCGGCAGCGUGUGGUCGCU-GUCGCCGAGUGUGGUUCGAAGGAU-AGCACGGAGA------------ ....((((((((((....)))))))..))).((((((((....)))((--(((((((((......))))-))))))).((((...((....)).-.)))))))))------------ ( -43.50, z-score = -2.52, R) >droSec1.super_64 139763 103 - 179039 AUUGGUCGGCUGUAUUUUUGCAGCUUUGGCAUCUCCCGUUACAGCGCGCGGCGGCAGCGUGUGGUCGCU-GUCGCCGAGUGUGGUUCGAAGGAU-AGCACGGAGA------------ ....((((((((((....)))))))..))).(((((.((((..((((.(((((((((((......))))-))))))).))))...((....)))-)))..)))))------------ ( -48.80, z-score = -3.15, R) >droAna3.scaffold_13250 984450 92 - 3535662 ----------UGUAUGUU-GUCACUUUAAGAAAAGUCGUUUUAGCAGG--GCGACAGUGUAGUGUCGCU-GUCACCGAGUGUGGUUCGGUGGUA-AGGAUAGAAAGA---------- ----------....((((-(.((((((....))))).)...))))).(--((((((......)))))))-.((((((((.....))))))))..-............---------- ( -23.70, z-score = -1.46, R) >droVir3.scaffold_12958 562607 111 + 3547706 ----GUCAGUGGUGGUAUACACACUUGGUGGUGUACACACGUAGUGGA--AUGUUGGCAUUGUAAAAUUAGUCUAAGAACAAAUCUCGUUGAUGCAACACGAUGUCACGCCCAAGGA ----....(((...((((((.(((...))))))))).)))....(((.--.(((.((((((((.......(((...((.......))...))).....))))))))))).))).... ( -25.30, z-score = 1.23, R) >droMoj3.scaffold_6498 2345717 96 + 3408170 ---CAAUAUUUGUAUUUU-GUCGCUUAAGAAG-UACCGUUUUAGCGGG--GCGACAGCGUUGUGUCGCU-GUCGCCGAGUGAGGUUCGAUGGUA-GGCACAAAGA------------ ---...........((((-((.(((((.(((.-(.((((....))))(--(((((((((......))))-))))))......).))).....))-))))))))).------------ ( -36.60, z-score = -3.60, R) >consensus ____GUCAGCUGUAUUUU_GUCGCUUUGGCAUCUACCGUUAUAGCGGG__GCGACAGCGUGGUGUCGCU_GUCGCCGAGUGUGGUUCGAUGGUA_AGCACGGAGA____________ .........((((.........(((.((((.......)))).))).....(((((((((......)))).))))).......................))))............... (-10.20 = -10.00 + -0.20)

| Location | 1,162,807 – 1,162,908 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 54.16 |
| Shannon entropy | 0.75989 |
| G+C content | 0.51081 |
| Mean single sequence MFE | -26.16 |
| Consensus MFE | -6.48 |
| Energy contribution | -6.28 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.67 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.25 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.953950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
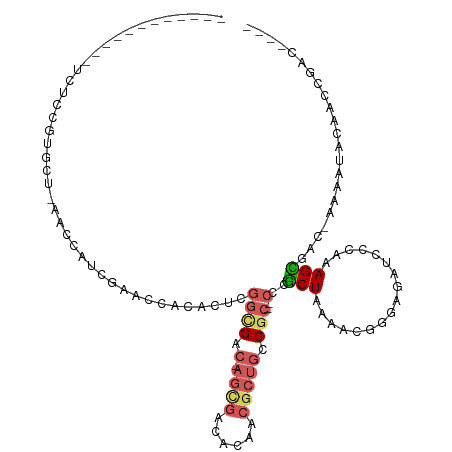
>dm3.chr3RHet 1162807 101 - 2517507 ------------UCUCCGUGCU-AUCCUUCGAACCACACUCGGCGAC-AGCGACCACACGCUGCCGC--CGCGCUGUAACGGGAGAUGCCAAAGCUACAAAAAUACAGCCGACCAAU ------------....((.(((-(((..(((....((((.(((((.(-((((......))))).)))--)).).)))..)))..)))((....))...........)))))...... ( -27.80, z-score = -1.62, R) >droSec1.super_64 139763 103 + 179039 ------------UCUCCGUGCU-AUCCUUCGAACCACACUCGGCGAC-AGCGACCACACGCUGCCGCCGCGCGCUGUAACGGGAGAUGCCAAAGCUGCAAAAAUACAGCCGACCAAU ------------(((((((((.-.......((.......))((((.(-((((......))))).))))))))(......).))))).......((((........))))........ ( -30.80, z-score = -1.37, R) >droAna3.scaffold_13250 984450 92 + 3535662 ----------UCUUUCUAUCCU-UACCACCGAACCACACUCGGUGAC-AGCGACACUACACUGUCGC--CCUGCUAAAACGACUUUUCUUAAAGUGAC-AACAUACA---------- ----------............-...((((((.......))))))..-.((((((......))))))--............(((((....)))))...-........---------- ( -18.60, z-score = -3.63, R) >droVir3.scaffold_12958 562607 111 - 3547706 UCCUUGGGCGUGACAUCGUGUUGCAUCAACGAGAUUUGUUCUUAGACUAAUUUUACAAUGCCAACAU--UCCACUACGUGUGUACACCACCAAGUGUGUAUACCACCACUGAC---- ((..((((((((.....(((((((((....((((((.(((....))).))))))...))).))))))--.....)))))(((((((((.....).))))))))..)))..)).---- ( -26.20, z-score = -0.75, R) >droMoj3.scaffold_6498 2345717 96 - 3408170 ------------UCUUUGUGCC-UACCAUCGAACCUCACUCGGCGAC-AGCGACACAACGCUGUCGC--CCCGCUAAAACGGUA-CUUCUUAAGCGAC-AAAAUACAAAUAUUG--- ------------..(((((((.-..................((((((-((((......)))))))))--)(((......)))..-........)).))-)))............--- ( -27.40, z-score = -4.46, R) >consensus ____________UCUCCGUGCU_AACCAUCGAACCACACUCGGCGAC_AGCGACACAACGCUGCCGC__CCCGCUAAAACGGGAGAUCCCAAAGCGAC_AAAAUACAACCGAC____ .........................................((((....(((((........)))))....)))).......................................... ( -6.48 = -6.28 + -0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:48:20 2011