
| Sequence ID | dm3.chr3RHet |
|---|---|
| Location | 1,075,502 – 1,075,612 |
| Length | 110 |
| Max. P | 0.500000 |

| Location | 1,075,502 – 1,075,612 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 53.89 |
| Shannon entropy | 0.84477 |
| G+C content | 0.39975 |
| Mean single sequence MFE | -22.62 |
| Consensus MFE | -7.43 |
| Energy contribution | -6.47 |
| Covariance contribution | -0.97 |
| Combinations/Pair | 1.55 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
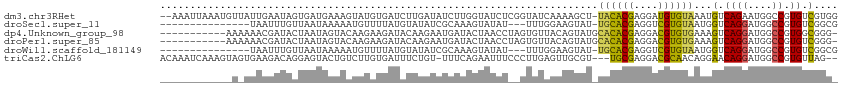
>dm3.chr3RHet 1075502 110 + 2517507 --AAAUUAAAUGUUAUUGAAUAGUGAUGAAAGUAUGUGAUCUUGAUAUCUUGGUAUCUCGGUAUCAAAAGCU-UACACGAGGAUGUGUAAAUGUCAGAAUGGCCGUGUCGUGG --.......................((((......((.((..((((((.(((((((....)))))))....(-(((((......))))))))))))..)).))....)))).. ( -19.00, z-score = 0.67, R) >droSec1.super_11 160440 94 + 2888827 ---------------UAAUUUGUUAAUAAAAAUGUUUUAUGUAUAUCGCAAAGUAUAU---UUUGGAAGUAU-UGCACGAGGUCGUGUAAUGGUCAGGAUGGCCGUGUCGGCG ---------------...(((((..(((((.....))))).......)))))...(((---(((((...(((-((((((....))))))))).))))))))((((...)))). ( -21.20, z-score = -1.17, R) >dp4.Unknown_group_98 10166 101 - 31687 -----------AAAAAACGAUACUAAUAGUACAAGAAGAUACAAGAAUGAUACUAACCUAGUGUUACAGUAUGCACACGAGGACGUGUGAAAGUCAGGAUGGCCGUGGCGGG- -----------........(((((....(((........))).....((((((((...)))))))).))))).((((((....))))))...((((((....)).))))...- ( -20.60, z-score = -0.78, R) >droPer1.super_85 10852 101 - 242291 -----------AAAAAACGAUACUAAUAGUACAAGAAGAUACAAGAAUGAUACUAACCUAGUGUUACAGUAUGCACACGAGGACGUGUGAAAGUCAGGAUGGCCGUGUCGGG- -----------......((((((..............(((.......((((((((...)))))))).......((((((....))))))...))).((....))))))))..- ( -22.80, z-score = -1.39, R) >droWil1.scaffold_181149 31542 94 - 589524 ---------------UAAUUUGUUAAUAAAAAUGUUUUAUGUAUAUCGCAAAGUAUAU---UUUGGAAGUAU-UGCACGAGGUCGUGUAAUGGUCAGGAUGGCCGUGUCGGCG ---------------...(((((..(((((.....))))).......)))))...(((---(((((...(((-((((((....))))))))).))))))))((((...)))). ( -21.20, z-score = -1.17, R) >triCas2.ChLG6 9481274 107 - 13544221 ACAAAUCAAAGUAGUGAAGACAGGAGUACUGUCUUGUGAUUUCUGU-UUUCAGAAUUUCCCUUGAGUUGCGU---UGCGAGGACGCAACAGGAACAGGAUGGCCGUGUUAG-- ........((((..(((((((((((((((......)).))))))))-)))))..)))).(((...(((((((---(.....)))))))))))((((((....)).))))..-- ( -30.90, z-score = -1.32, R) >consensus ____________A_AUAAGAUACUAAUAAAAAUAUAUGAUAUAUAUAUCAUAGUAUCU__GUUUGAAAGUAU_UACACGAGGACGUGUAAAGGUCAGGAUGGCCGUGUCGGG_ .........................................................................((((((....))))))...(.((((....)).)).).... ( -7.43 = -6.47 + -0.97)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:48:16 2011