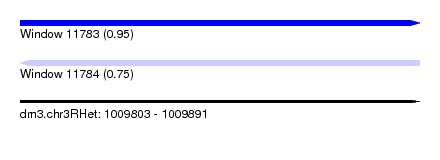
| Sequence ID | dm3.chr3RHet |
|---|---|
| Location | 1,009,803 – 1,009,891 |
| Length | 88 |
| Max. P | 0.953965 |

| Location | 1,009,803 – 1,009,891 |
|---|---|
| Length | 88 |
| Sequences | 5 |
| Columns | 88 |
| Reading direction | forward |
| Mean pairwise identity | 74.09 |
| Shannon entropy | 0.47485 |
| G+C content | 0.54188 |
| Mean single sequence MFE | -24.58 |
| Consensus MFE | -14.80 |
| Energy contribution | -17.08 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.953965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
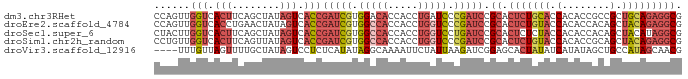
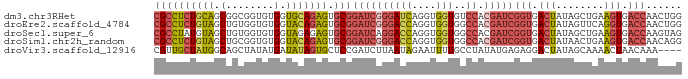
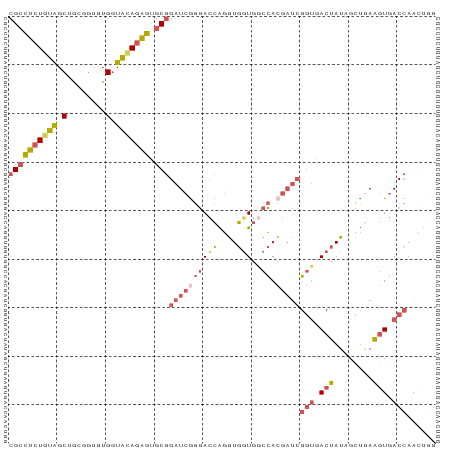
>dm3.chr3RHet 1009803 88 + 2517507 CCAGUUGGUCACUUCAGCUAUAGUCACCGAUCGUGGACACCACCUGAUCCCGAUCCGCACUCUGCACCACACCGCCGCUGCAGAGGCG ......(((.(((........))).)))(((((.(((((.....)).))))))))(((.(((((((.(........).)))))))))) ( -29.30, z-score = -2.55, R) >droEre2.scaffold_4784 24019961 88 + 25762168 CCAGUUGGUCACCUGAACUAUAGUCACCGAUCGUGGCCACCACCUGGUCCCGAUCCGCACUCUGUACCACACCACAGCUACAGAGGCG ......(((.((..........)).)))(((((.(((((.....))))).)))))(((.(((((((.(........).)))))))))) ( -27.70, z-score = -1.97, R) >droSec1.super_6 17490 88 + 4358794 CUACUUGGUCACUUCAGCUAUAGUCACCGAUCGUGGCCACCACCUGGUCCUGAUCCGCACUCUCUACCACACCACAGCUACAUAGGCG ......(((.(((........))).)))(((((.(((((.....))))).))))).....................(((.....))). ( -18.60, z-score = -0.52, R) >droSim1.chr2h_random 1477450 88 + 3178526 CCUGUUGGUCACUUCAGUUAUAGUCACCGAUCGUGGCCACCACCUGGUCCCGAUCCGCACUCUGUACCACACCGCAGCUACAGAGGCG ......(((.(((........))).)))(((((.(((((.....))))).)))))(((.(((((((.(........).)))))))))) ( -30.50, z-score = -2.79, R) >droVir3.scaffold_12916 38087 84 + 114074 ----UUUGUUAGUUUUGCUAUAGUCCUCUCAUAUAGGCAAAAUUCUAUUAAGAUCGGAGCACUAUAUCAUAUAGCUGCCAUAGCAACG ----..........(((((((..(((..((..(((((......)))))...))..)))((((((((...))))).))).))))))).. ( -16.80, z-score = -1.23, R) >consensus CCAGUUGGUCACUUCAGCUAUAGUCACCGAUCGUGGCCACCACCUGGUCCCGAUCCGCACUCUGUACCACACCGCAGCUACAGAGGCG ......(((.(((........))).)))(((((.(((((.....))))).))))).((.(((((((.(........).))))))))). (-14.80 = -17.08 + 2.28)
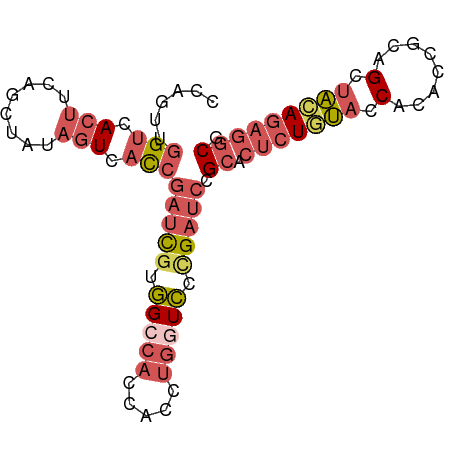
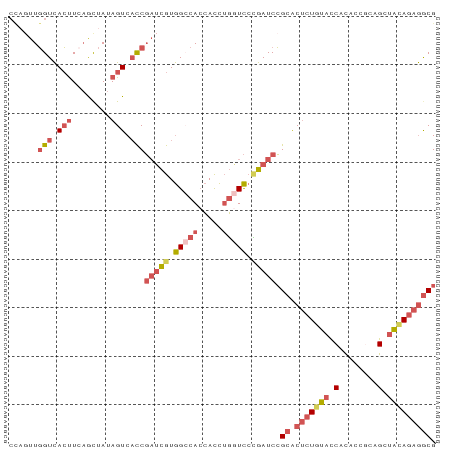
| Location | 1,009,803 – 1,009,891 |
|---|---|
| Length | 88 |
| Sequences | 5 |
| Columns | 88 |
| Reading direction | reverse |
| Mean pairwise identity | 74.09 |
| Shannon entropy | 0.47485 |
| G+C content | 0.54188 |
| Mean single sequence MFE | -30.00 |
| Consensus MFE | -18.26 |
| Energy contribution | -19.86 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.749671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
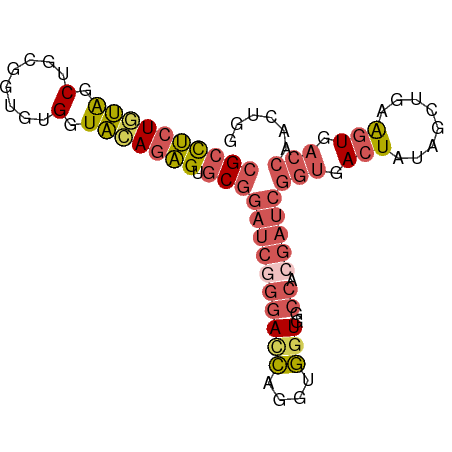
>dm3.chr3RHet 1009803 88 - 2517507 CGCCUCUGCAGCGGCGGUGUGGUGCAGAGUGCGGAUCGGGAUCAGGUGGUGUCCACGAUCGGUGACUAUAGCUGAAGUGACCAACUGG ((((((((((.((......)).))))))).)))(((((((((........)))).)))))(((.(((........))).)))...... ( -36.20, z-score = -1.63, R) >droEre2.scaffold_4784 24019961 88 - 25762168 CGCCUCUGUAGCUGUGGUGUGGUACAGAGUGCGGAUCGGGACCAGGUGGUGGCCACGAUCGGUGACUAUAGUUCAGGUGACCAACUGG ((((((((((.(........).))))))).)))(((((((.(((.....))))).)))))(((.(((........))).)))...... ( -35.70, z-score = -1.61, R) >droSec1.super_6 17490 88 - 4358794 CGCCUAUGUAGCUGUGGUGUGGUAGAGAGUGCGGAUCAGGACCAGGUGGUGGCCACGAUCGGUGACUAUAGCUGAAGUGACCAAGUAG (((.....((((((((((...........(((.((((.((.(((.....)))))..)))).)))))))))))))..)))......... ( -22.20, z-score = 1.06, R) >droSim1.chr2h_random 1477450 88 - 3178526 CGCCUCUGUAGCUGCGGUGUGGUACAGAGUGCGGAUCGGGACCAGGUGGUGGCCACGAUCGGUGACUAUAACUGAAGUGACCAACAGG ((((((((((.(........).))))))).)))(((((((.(((.....))))).)))))(((.(((........))).)))...... ( -35.70, z-score = -2.05, R) >droVir3.scaffold_12916 38087 84 - 114074 CGUUGCUAUGGCAGCUAUAUGAUAUAGUGCUCCGAUCUUAAUAGAAUUUUGCCUAUAUGAGAGGACUAUAGCAAAACUAACAAA---- ..(((((((((..((((((...))))))..(((..(((((((((........)))).)))))))))))))))))..........---- ( -20.20, z-score = -1.98, R) >consensus CGCCUCUGUAGCUGCGGUGUGGUACAGAGUGCGGAUCGGGACCAGGUGGUGGCCACGAUCGGUGACUAUAGCUGAAGUGACCAACUGG ((((((((((.(........).))))))).)))((((((((((....)))..)).)))))(((.(((........))).)))...... (-18.26 = -19.86 + 1.60)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:48:13 2011