| Sequence ID | dm3.chr3RHet |
|---|---|
| Location | 1,006,061 – 1,006,190 |
| Length | 129 |
| Max. P | 0.961735 |

| Location | 1,006,061 – 1,006,190 |
|---|---|
| Length | 129 |
| Sequences | 6 |
| Columns | 131 |
| Reading direction | forward |
| Mean pairwise identity | 70.91 |
| Shannon entropy | 0.56567 |
| G+C content | 0.54395 |
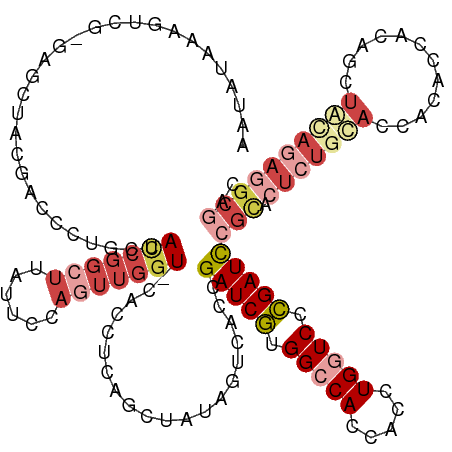
| Mean single sequence MFE | -39.73 |
| Consensus MFE | -15.95 |
| Energy contribution | -19.22 |
| Covariance contribution | 3.27 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.961735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
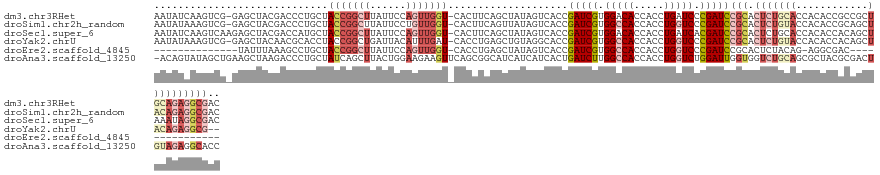
>dm3.chr3RHet 1006061 129 + 2517507 AAUAUCAAGUCG-GAGCUACGACCCUGCUACCGGCUUAUUCCAGUUGGU-CACUUCAGCUAUAGUCACCGAUCGUGGACACCACCUGAUCCCGAUCCGCACUCUGCACCACACCGCCGCUGCAGAGGCGAC ......((((((-((((.........))).))))))).....((((((.-....))))))...(((...(((((.(((((.....)).)))))))).((.(((((((.(........).)))))))))))) ( -43.50, z-score = -3.10, R) >droSim1.chr2h_random 1477411 129 + 3178526 AAUAUAAAGUCG-GAGCUACGACCCUGCUACCGGCUUAUUCCUGUUGGU-CACUUCAGUUAUAGUCACCGAUCGUGGCCACCACCUGGUCCCGAUCCGCACUCUGUACCACACCGCAGCUACAGAGGCGAC ......((((((-((((.........))).))))))).........(((-.(((........))).)))(((((.(((((.....))))).)))))(((.(((((((.(........).)))))))))).. ( -44.50, z-score = -3.16, R) >droSec1.super_6 16741 130 + 4358794 AAUAUCAAGUCAAGAGCUACGACCAUGCUACCGGCUUAUUCCAGUUGGU-CACUUCAGCUAUAGUCACCGAUCGUGGACACCACCUGAUCACGAUCCGCACUCUGCACCACACCACAGCUAAAUAGGCGAC ........(((.((((....(((.((((((((((((......)))))))-......))).)).)))...((((((((.((.....)).))))))))....)))).............(((.....)))))) ( -30.40, z-score = -1.08, R) >droYak2.chrU 1895624 127 + 28119190 AAUAUAAAGUCG-GAGCUACAACGCACCUACCGGCUGAUUACAUUUGAU-CACCUGAGCUGUAGGCACCGAUCGUGGCCACCACCUGGUCCCGAUCCGCACUCUGUACCACACCACAGCUACAGAGGCG-- ...........(-(.((......)).((((((((.((((((....))))-)).)))....)))))..))(((((.(((((.....))))).)))))(((.(((((((.(........).))))))))))-- ( -40.20, z-score = -2.50, R) >droEre2.scaffold_4845 20484219 100 - 22589142 --------------UAUUUAAAGCCUGCUACCGGCUUAUUCCAGUUGGU-CACCUGAGCUAUAGUCACCGAUCGUGGCCACCACCUGGUCCCGAUCCGCACUCUACAG-AGGCGAC--------------- --------------.......((((.(..(((((((......)))))))-..)..).)))...(((...(((((.(((((.....))))).))))).((.(((....)-)))))))--------------- ( -29.90, z-score = -1.56, R) >droAna3.scaffold_13250 1019668 130 + 3535662 -ACAGUAUAGCUGAAGCUAAGACCCUGCUAUCAGCUUACUGGAAGAAGUUCAGCGGCAUCAUCAUCACUGAUCUUGGCCACCACCUGGUCUGGAUUGGUGGUCUGCAGCGCUACGCGACUGUAGAGGCACC -.(((((.(((((((((.........))).)))))))))))...........((..........((((..((((.(((((.....))))).))))..))))((((((((((...))).))))))).))... ( -49.90, z-score = -2.28, R) >consensus AAUAUAAAGUCG_GAGCUACGACCCUGCUACCGGCUUAUUCCAGUUGGU_CACCUCAGCUAUAGUCACCGAUCGUGGCCACCACCUGGUCCCGAUCCGCACUCUGCACCACACCACAGCUACAGAGGCGAC .............................(((((((......)))))))....................(((((.(((((.....))))).)))))(((.(((((((.(........).)))))))))).. (-15.95 = -19.22 + 3.27)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:48:11 2011