| Sequence ID | dm3.chr3RHet |
|---|---|
| Location | 918,734 – 918,799 |
| Length | 65 |
| Max. P | 0.995283 |

| Location | 918,734 – 918,799 |
|---|---|
| Length | 65 |
| Sequences | 4 |
| Columns | 67 |
| Reading direction | reverse |
| Mean pairwise identity | 65.08 |
| Shannon entropy | 0.56504 |
| G+C content | 0.44667 |
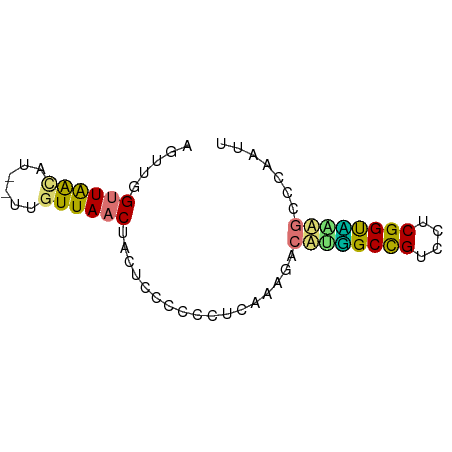
| Mean single sequence MFE | -14.25 |
| Consensus MFE | -11.14 |
| Energy contribution | -10.33 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.79 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.995283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
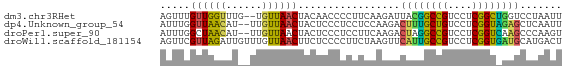
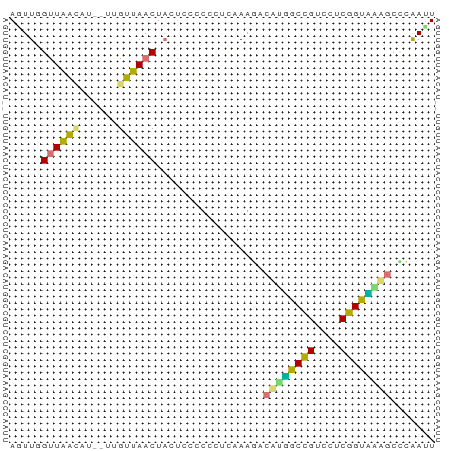
>dm3.chr3RHet 918734 65 - 2517507 AGUUUGUUGGUUUG--UGUUAACUACAACCCCUUCAAGAUUACGGCCGUCCUCGGCUGGUCCUAAUU ..((((..((.(((--((.....))))).))...))))....((((((....))))))......... ( -15.90, z-score = -1.31, R) >dp4.Unknown_group_54 47496 65 - 61718 AUUUGGUUAACAU--UUGUUAACUACUCCCUCCUCCAAGACUUUGCUGUCCUCGGUAGAGCUCAAUU ...((((((((..--..)))))))).............((((((((((....)))))))).)).... ( -16.60, z-score = -3.40, R) >droPer1.super_90 61284 65 - 211371 AUUUGGCUAACAU--UUGUUAACUACUCCCUCCUUCAAGACUAGGCCGUCCUCGGUCAAGCCCAAGU ....(((((((..--..))))......................(((((....)))))..)))..... ( -12.80, z-score = -1.05, R) >droWil1.scaffold_181154 1377996 67 - 4610121 AGUUCGUUAGAUUGUUUGUUAACUUCUCCCCUUCUAAGUUCAUUGCCGUCCUCGGUGAUGCAUGACU .....(..(((..(((....))).)))..)......(((((((..(((....)))..)))...)))) ( -11.70, z-score = -1.21, R) >consensus AGUUGGUUAACAU__UUGUUAACUACUCCCCCCUCAAAGACAUGGCCGUCCUCGGUAAAGCCCAAUU .....((((((......)))))).................((((((((....))))))))....... (-11.14 = -10.33 + -0.81)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:48:07 2011