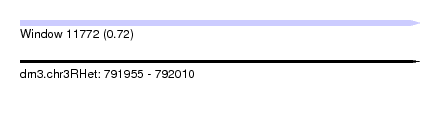
| Sequence ID | dm3.chr3RHet |
|---|---|
| Location | 791,955 – 792,010 |
| Length | 55 |
| Max. P | 0.721558 |

| Location | 791,955 – 792,010 |
|---|---|
| Length | 55 |
| Sequences | 4 |
| Columns | 66 |
| Reading direction | forward |
| Mean pairwise identity | 60.58 |
| Shannon entropy | 0.62177 |
| G+C content | 0.65061 |
| Mean single sequence MFE | -21.20 |
| Consensus MFE | -13.43 |
| Energy contribution | -12.80 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.71 |
| Mean z-score | -0.42 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.721558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
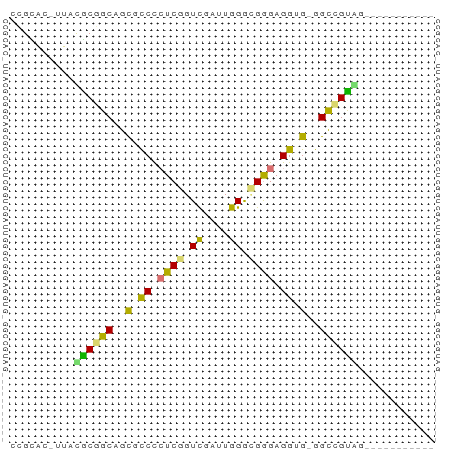
>dm3.chr3RHet 791955 55 + 2517507 CCGUACGAUACACGGCAGCCCCCCUCGGUCGGUUGGGCGGGAGGUGUGGCCGUGG----------- ..........((((((.(((((((((((....))))).))).)))...)))))).----------- ( -25.70, z-score = -0.75, R) >droPer1.super_57 61412 61 - 429542 --GCGCAUCAUGCGGCAGUGCCCAUCGGUCGAUUGGGGGGGAGGAG-GGCUGCGUUUUUCCUGG-- --.(((.....)))......((((((....)).))))((((((((.-(....).))))))))..-- ( -17.80, z-score = 1.34, R) >droAna3.scaffold_13499 582714 62 - 1739075 CCGUA---UACGCGGCAGCGUCUCUCGGUCGAUUGGACGGGAGGUG-GGCUGUAUGUAUACAGUGG ..(((---((((((((..(..((((((..(....)..))))))..)-.)))))..))))))..... ( -25.30, z-score = -2.52, R) >droGri2.scaffold_15110 22582108 53 + 24565398 CUGCACUUUAUGCAGC-GCGCCCCUCGGUUGGUUGGGUGAGAGGUG-GGUCGUAG----------- ..........(((.((-.((((.((((.((....)).)))).))))-.)).))).----------- ( -16.00, z-score = 0.26, R) >consensus CCGCAC_UUACGCGGCAGCGCCCCUCGGUCGAUUGGGCGGGAGGUG_GGCCGUAG___________ ..........((((((....((.((((.((....)).)))).))....))))))............ (-13.43 = -12.80 + -0.63)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:48:03 2011