| Sequence ID | dm3.chr3RHet |
|---|---|
| Location | 759,520 – 759,616 |
| Length | 96 |
| Max. P | 0.898914 |

| Location | 759,520 – 759,616 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 52.84 |
| Shannon entropy | 0.66421 |
| G+C content | 0.49774 |
| Mean single sequence MFE | -17.97 |
| Consensus MFE | -11.66 |
| Energy contribution | -9.68 |
| Covariance contribution | -1.98 |
| Combinations/Pair | 1.67 |
| Mean z-score | -0.31 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.810128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3RHet 759520 96 + 2517507 GCCCUACCCUACCUAAGACCACUUUCCUUAAGAGCAUGGCUCUAGUGGUCCCUUUCUGCACCCUACGUCCGGAAAAACAUCAGCCGAGUCAGUAUA ((..............(((((((.......(((((...))))))))))))..((((((.((.....)).)))))).......))............ ( -19.11, z-score = -0.50, R) >droYak2.chr2L_random 1704470 78 - 4064425 -------------UACGAUACCCUGCUGCUAGAGCAUGGCUUUGGUGAUCCCAUUGAUCACCCUACGACAGCGAUAAUAUCAGUUAAGUUA----- -------------...((((...((((((.(((((...)))))(((((((.....)))))))....).)))))....))))..........----- ( -19.80, z-score = -1.95, R) >droAna3.scaffold_12928 120838 88 - 771024 ----AACCCUACC-AUGGCCACAGGCAG-AAGAGUGUGACUCUCGUGGCCCCUUUUCCCCCGUUACACCCGAGGAUACAUUUGCGAAGUUAGUU-- ----....(((.(-..((((((.....(-(.(((.....))))))))))).......((.((.......)).)).............).)))..-- ( -15.00, z-score = 1.51, R) >consensus _____ACCCUACCUAAGACCACAUGCAG_AAGAGCAUGGCUCUAGUGGUCCCUUUCACCACCCUACGACCGAGAAAACAUCAGCCAAGUUAGU___ ................(((((((.......(((((...))))))))))))..((((((...........))))))..................... (-11.66 = -9.68 + -1.98)


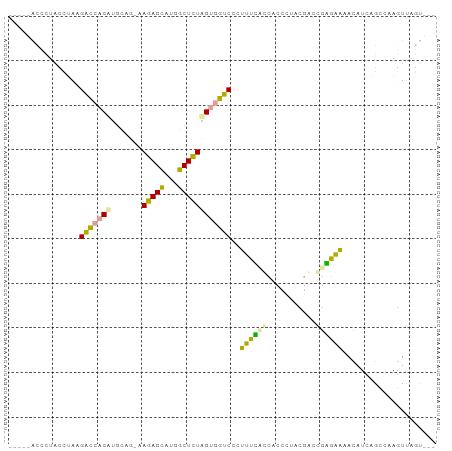
| Location | 759,520 – 759,616 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 52.84 |
| Shannon entropy | 0.66421 |
| G+C content | 0.49774 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -14.11 |
| Energy contribution | -12.68 |
| Covariance contribution | -1.43 |
| Combinations/Pair | 1.61 |
| Mean z-score | -0.88 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.898914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3RHet 759520 96 - 2517507 UAUACUGACUCGGCUGAUGUUUUUCCGGACGUAGGGUGCAGAAAGGGACCACUAGAGCCAUGCUCUUAAGGAAAGUGGUCUUAGGUAGGGUAGGGC ....((.((((.(((..(((...(((.......))).)))....((((((((((((((...))))).......))))))))).))).)))).)).. ( -27.11, z-score = 0.04, R) >droYak2.chr2L_random 1704470 78 + 4064425 -----UAACUUAACUGAUAUUAUCGCUGUCGUAGGGUGAUCAAUGGGAUCACCAAAGCCAUGCUCUAGCAGCAGGGUAUCGUA------------- -----..........((((((...(((((..((((((((((.....)))))))..(((...)))))))))))..))))))...------------- ( -23.70, z-score = -2.07, R) >droAna3.scaffold_12928 120838 88 + 771024 --AACUAACUUCGCAAAUGUAUCCUCGGGUGUAACGGGGGAAAAGGGGCCACGAGAGUCACACUCUU-CUGCCUGUGGCCAU-GGUAGGGUU---- --....((((..((.......(((((.(......).))))).....(((((((((((.....)))))-......))))))..-.))..))))---- ( -28.10, z-score = -0.60, R) >consensus ___ACUAACUUAGCUGAUGUUUUCCCGGACGUAGGGUGAAAAAAGGGACCACCAGAGCCAUGCUCUU_CAGCAAGUGGUCAUAGGUAGGGU_____ .....................((((((.......))))))......((((((((((((...))))).......)))))))................ (-14.11 = -12.68 + -1.43)

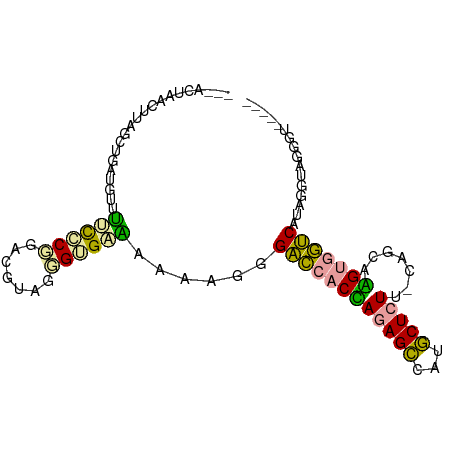

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:48:02 2011